Combinatorial strategies targeting NEAT1 and AURKA as new potential therapeutic options for multiple myeloma
- PMID: 38988264
- PMCID: PMC11609815
- DOI: 10.3324/haematol.2024.285470
Combinatorial strategies targeting NEAT1 and AURKA as new potential therapeutic options for multiple myeloma
Abstract
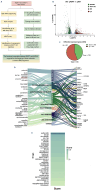
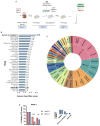
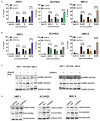
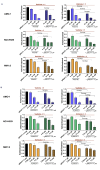
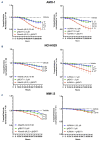
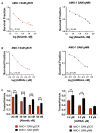
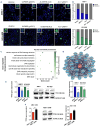
Multiple myeloma (MM) is a dreadful disease, marked by the uncontrolled proliferation of clonal plasma cells within the bone marrow. It is characterized by a highly heterogeneous clinical and molecular background, supported by severe genomic alterations. Important de-regulation of long non-coding RNA (lncRNA) expression, which can influence progression and therapy resistance, has been reported in MM patients. NEAT1 is a lncRNA essential for nuclear paraspeckles and is involved in the regulation of gene expression. We showed that NEAT1 supports MM proliferation, making this lncRNA an attractive therapeutic candidate. Here, we used a combinatorial strategy integrating transcriptomic and computational approaches with functional high-throughput drug screening to identify compounds that synergize with NEAT1 inhibition in restraining MM cell growth. AURKA inhibitors were identified as top-scoring drugs in these analyses. We showed that the combination of NEAT1 silencing and AURKA inhibitors in MM profoundly impairs microtubule organization and mitotic spindle assembly, finally leading to cell death. Analysis of the large publicly available CoMMpass dataset showed that, in MM patients, AURKA expression is strongly associated with reduced progression-free survival (P<0.0001) and overall survival (P<0.0001) probabilities and patients with high levels of expression of both NEAT1 and AURKA have a worse clinical outcome. Finally, using RNA-sequencing data from NEAT1 knockdown MM cells, we identified the AURKA allosteric regulator TPX2 as a new NEAT1 target in MM and as a mediator of the interplay between AURKA and NEAT1, therefore providing a possible explanation for the synergistic activity observed upon their combinatorial inhibition.
Figures
References
-
- Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364(11):1046-1060. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cance J Clin 2016;66(1):7-30. - PubMed
-
- Rajkumar SV. Multiple myeloma: 2020 update on diagnosis, risk-stratification and management. Am J Hematol. 2020;95(5):548-567. - PubMed
-
- Quinn JJ, Chang H. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47-62. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

