Impact of tRNA-induced proline-to-serine mistranslation on the transcriptome of Drosophila melanogaster
- PMID: 38989890
- PMCID: PMC11373654
- DOI: 10.1093/g3journal/jkae151
Impact of tRNA-induced proline-to-serine mistranslation on the transcriptome of Drosophila melanogaster
Abstract
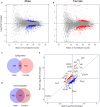
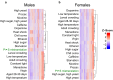
Mistranslation is the misincorporation of an amino acid into a polypeptide. Mistranslation has diverse effects on multicellular eukaryotes and is implicated in several human diseases. In Drosophila melanogaster, a serine transfer RNA (tRNA) that misincorporates serine at proline codons (P→S) affects male and female flies differently. The mechanisms behind this discrepancy are currently unknown. Here, we compare the transcriptional response of male and female flies to P→S mistranslation to identify genes and cellular processes that underlie sex-specific differences. Both males and females downregulate genes associated with various metabolic processes in response to P→S mistranslation. Males downregulate genes associated with extracellular matrix organization and response to negative stimuli such as wounding, whereas females downregulate aerobic respiration and ATP synthesis genes. Both sexes upregulate genes associated with gametogenesis, but females also upregulate cell cycle and DNA repair genes. These observed differences in the transcriptional response of male and female flies to P→S mistranslation have important implications for the sex-specific impact of mistranslation on disease and tRNA therapeutics.
Keywords: Drosophila melanogaster; RNA sequencing; mistranslation; proteotoxicity; stress response; tRNA; translation.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The authors declare no conflicts of interest.
Figures


Update of
-
Impact of tRNA-induced proline-to-serine mistranslation on the transcriptome of Drosophila melanogaster.bioRxiv [Preprint]. 2024 May 10:2024.05.08.593249. doi: 10.1101/2024.05.08.593249. bioRxiv. 2024. Update in: G3 (Bethesda). 2024 Sep 4;14(9):jkae151. doi: 10.1093/g3journal/jkae151. PMID: 38766246 Free PMC article. Updated. Preprint.
Similar articles
-
Impact of tRNA-induced proline-to-serine mistranslation on the transcriptome of Drosophila melanogaster.bioRxiv [Preprint]. 2024 May 10:2024.05.08.593249. doi: 10.1101/2024.05.08.593249. bioRxiv. 2024. Update in: G3 (Bethesda). 2024 Sep 4;14(9):jkae151. doi: 10.1093/g3journal/jkae151. PMID: 38766246 Free PMC article. Updated. Preprint.
-
Mistranslating tRNA variants have anticodon- and sex-specific impacts on Drosophila melanogaster.bioRxiv [Preprint]. 2024 Jun 13:2024.06.11.598535. doi: 10.1101/2024.06.11.598535. bioRxiv. 2024. Update in: G3 (Bethesda). 2024 Sep 23:jkae230. doi: 10.1093/g3journal/jkae230. PMID: 38915589 Free PMC article. Updated. Preprint.
-
Mistranslating tRNA variants have anticodon- and sex-specific impacts on Drosophila melanogaster.G3 (Bethesda). 2024 Sep 23;14(12):jkae230. doi: 10.1093/g3journal/jkae230. Online ahead of print. G3 (Bethesda). 2024. PMID: 39312260 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Allen E. 2016. RNA extraction from Drosophila tissues using TRIzol reagent. protocols.io. doi: 10.17504/protocols.io.fgtbjwn. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
