Following the Evolutionary Paths of Dscam1 Proteins toward Highly Specific Homophilic Interactions
- PMID: 38989909
- PMCID: PMC11272049
- DOI: 10.1093/molbev/msae141
Following the Evolutionary Paths of Dscam1 Proteins toward Highly Specific Homophilic Interactions
Abstract
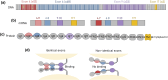
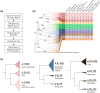
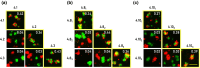
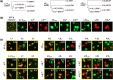
Many adhesion proteins, evolutionarily related through gene duplication, exhibit distinct and precise interaction preferences and affinities crucial for cell patterning. Yet, the evolutionary paths by which these proteins acquire new specificities and prevent cross-interactions within their family members remain unknown. To bridge this gap, this study focuses on Drosophila Down syndrome cell adhesion molecule-1 (Dscam1) proteins, which are cell adhesion proteins that have undergone extensive gene duplication. Dscam1 evolved under strong selective pressure to achieve strict homophilic recognition, essential for neuronal self-avoidance and patterning. Through a combination of phylogenetic analyses, ancestral sequence reconstruction, and cell aggregation assays, we studied the evolutionary trajectory of Dscam1 exon 4 across various insect lineages. We demonstrated that recent Dscam1 duplications in the mosquito lineage bind with strict homophilic specificities without any cross-interactions. We found that ancestral and intermediate Dscam1 isoforms maintained their homophilic binding capabilities, with some intermediate isoforms also engaging in promiscuous interactions with other paralogs. Our results highlight the robust selective pressure for homophilic specificity integral to the Dscam1 function within the process of neuronal self-avoidance. Importantly, our study suggests that the path to achieving such selective specificity does not introduce disruptive mutations that prevent self-binding but includes evolutionary intermediates that demonstrate promiscuous heterophilic interactions. Overall, these results offer insights into evolutionary strategies that underlie adhesion protein interaction specificities.
Keywords: adhesion proteins; cell-cell recognition; down syndrome cell adhesion molecule, DSCAM; gene duplication; protein evolution; protein–protein interactions.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

