Genomic analysis and identification of a novel superantigen, SargEY, in Staphylococcus argenteus isolated from atopic dermatitis lesions
- PMID: 38990001
- PMCID: PMC11288046
- DOI: 10.1128/msphere.00505-24
Genomic analysis and identification of a novel superantigen, SargEY, in Staphylococcus argenteus isolated from atopic dermatitis lesions
Abstract
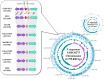
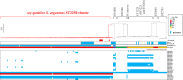
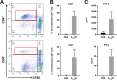
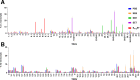
During surveillance of Staphylococcus aureus in lesions from patients with atopic dermatitis (AD), we isolated Staphylococcus argenteus, a species registered in 2011 as a new member of the genus Staphylococcus and previously considered a lineage of S. aureus. Genome sequence comparisons between S. argenteus isolates and representative S. aureus clinical isolates from various origins revealed that the S. argenteus genome from AD patients closely resembles that of S. aureus causing skin infections. We previously reported that 17%-22% of S. aureus isolated from skin infections produce staphylococcal enterotoxin Y (SEY), which predominantly induces T-cell proliferation via the T-cell receptor (TCR) Vα pathway. Complete genome sequencing of S. argenteus isolates revealed a gene encoding a protein similar to superantigen SEY, designated as SargEY, on its chromosome. Population structure analysis of S. argenteus revealed that these isolates are ST2250 lineage, which was the only lineage positive for the SEY-like gene among S. argenteus. Recombinant SargEY demonstrated immunological cross-reactivity with anti-SEY serum. SargEY could induce proliferation of human CD4+ and CD8+ T cells, as well as production of TNF-α and IFN-γ. SargEY showed emetic activity in a marmoset monkey model. SargEY and SET (a phylogenetically close but uncharacterized SE) revealed their dependency on TCR Vα in inducing human T-cell proliferation. Additionally, TCR sequencing revealed other previously undescribed Vα repertoires induced by SEH. SargEY and SEY may play roles in exacerbating the respective toxin-producing strains in AD.
Importance: Staphylococcus aureus is frequently isolated from active lesions of atopic dermatitis (AD) patients. We reported that 17%-22% of S. aureus isolated from AD patients produced a novel superantigen staphylococcal enterotoxin Y (SEY). Unlike many S. aureus superantigens that activate T cells via T-cell receptor (TCR) Vß, SEY activates T cells via TCR Vα and stimulates cytokine secretion. Staphylococcus argenteus was isolated from AD patients during the surveillance for S. aureus. Phylogenetic comparison of the genome indicated that the isolate was very similar to S. aureus causing skin infections. The isolate encoded a SEY-like protein, designated SargEY, which, like SEY, activated T cells via the TCR Vα. ST2250 is the only lineage positive for SargEY gene. ST2250 S. argenteus harboring a superantigen SargEY gene may be a novel staphylococcal clone that infects human skin and is involved in the exacerbation of AD.
Keywords: Staphylococcus; TCR Vα; argenteus; enterotoxins; human T-cell; superantigen.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Moradigaravand D, Jamrozy D, Mostowy R, Anderson A, Nickerson EK, Thaipadungpanit J, Wuthiekanun V, Limmathurotsakul D, Tandhavanant S, Wikraiphat C, Wongsuvan G, Teerawattanasook N, Jutrakul Y, Srisurat N, Chaimanee P, Eoin West T, Blane B, Parkhill J, Chantratita N, Peacock SJ. 2017. Evolution of the Staphylococcus argenteus ST2250 clone in northeastern Thailand is linked with the acquisition of livestock-associated staphylococcal genes. mBio 8:e00802-17. doi:10.1128/mBio.00802-17 - DOI - PMC - PubMed
-
- Tong SYC, Schaumburg F, Ellington MJ, Corander J, Pichon B, Leendertz F, Bentley SD, Parkhill J, Holt DC, Peters G, Giffard PM. 2015. Novel staphylococcal species that form part of a Staphylococcus aureus-related complex: the non-pigmented Staphylococcus argenteus sp. nov. and the non-human primate-associated Staphylococcus schweitzeri sp. nov. Int J Syst Evol Microbiol 65:15–22. doi:10.1099/ijs.0.062752-0 - DOI - PMC - PubMed
-
- Aung MS, San T, Aye MM, Mya S, Maw WW, Zan KN, Htut WHW, Kawaguchiya M, Urushibara N, Kobayashi N. 2017. Prevalence and genetic characteristics of Staphylococcus aureus and Staphylococcus argenteus isolates harboring panton-valentine leukocidin, enterotoxins, and TSST-1 genes from food handlers in Myanmar. Toxins (Basel) 9:241. doi:10.3390/toxins9080241 - DOI - PMC - PubMed
-
- Wakabayashi Y, Takemoto K, Iwasaki S, Yajima T, Kido A, Yamauchi A, Kuroiwa K, Kumai Y, Yoshihara S, Tokumoto H, Kawatsu K, Yasugi M, Miyake M. 2022. Isolation and characterization of Staphylococcus argenteus strains from retail foods and slaughterhouses in Japan. Int J Food Microbiol 363:109503. doi:10.1016/j.ijfoodmicro.2021.109503 - DOI - PubMed
MeSH terms
Substances
Grants and funding
- JP20gm1010001/Japan Agency for Medical Research and Development (AMED)
- 18ek0410028h0003/Japan Agency for Medical Research and Development (AMED)
- 21ek0410058h0003/Japan Agency for Medical Research and Development (AMED)
- 21gm1010001h0006/Japan Agency for Medical Research and Development (AMED)
- 24ek0410098h0003/Japan Agency for Medical Research and Development (AMED)
LinkOut - more resources
Full Text Sources
Research Materials
