Crystal structure of the class A extended-spectrum β-lactamase CTX-M-96 in complex with relebactam at 1.03 Angstrom resolution
- PMID: 38990013
- PMCID: PMC11304709
- DOI: 10.1128/aac.01721-23
Crystal structure of the class A extended-spectrum β-lactamase CTX-M-96 in complex with relebactam at 1.03 Angstrom resolution
Abstract
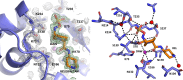
The use of β-lactam/β-lactamase inhibitors constitutes an important strategy to counteract β-lactamases in multidrug-resistant (MDR) Gram-negative bacteria. Recent reports have described ceftazidime-/avibactam-resistant isolates producing CTX-M variants with different amino acid substitutions (e.g., P167S, L169Q, and S130G). Relebactam (REL) combined with imipenem has proved very effective against Enterobacterales producing ESBLs, serine-carbapenemases, and AmpCs. Herein, we evaluated the inhibitory efficacy of REL against CTX-M-96, a CTX-M-15-type variant. The CTX-M-96 structure was obtained in complex with REL at 1.03 Å resolution (PDB 8EHH). REL was covalently bound to the S70-Oγ atom upon cleavage of the C7-N6 bond. Compared with apo CTX-M-96, binding of REL forces a slight displacement of the deacylating water inwards the active site (0.81 Å), making the E166 and N170 side chains shift to create a proper hydrogen bonding network. Binding of REL also disturbs the hydrophobic patch formed by Y105, P107, and Y129, likely due to the piperidine ring of REL that creates clashes with these residues. Also, a remarkable change in the positioning of the N104 sidechain is also affected by the piperidine ring. Therefore, differences in the kinetic behavior of REL against class A β-lactamases seem to rely, at least in part, on differences in the residues being involved in the association and stabilization of the inhibitor before hydrolysis. Our data provide the biochemical and structural basis for REL effectiveness against CTX-M-producing Gram-negative pathogens and essential details for further DBO design. Imipenem/REL remains an important choice for dealing with isolates co-producing CTX-M with other β-lactamases.
Keywords: 1,4-diazabicyclooctane; DBO; ESBL; X-ray crystallography; ceftazidimase; enzyme kinetics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Molecular Basis of Class A β-Lactamase Inhibition by Relebactam.Antimicrob Agents Chemother. 2019 Sep 23;63(10):e00564-19. doi: 10.1128/AAC.00564-19. Print 2019 Oct. Antimicrob Agents Chemother. 2019. PMID: 31383664 Free PMC article.
-
Rapid emergence of resistance to broad-spectrum direct antimicrobial activity of avibactam.Microbiol Spectr. 2025 Aug 5;13(8):e0324124. doi: 10.1128/spectrum.03241-24. Epub 2025 Jun 12. Microbiol Spectr. 2025. PMID: 40503840 Free PMC article.
-
Impact of the double deletion ΔG242-T243 in KPC-2 in the effectiveness of ceftazidime-avibactam and imipenem-relebactam.Antimicrob Agents Chemother. 2025 Jun 4;69(6):e0191524. doi: 10.1128/aac.01915-24. Epub 2025 May 5. Antimicrob Agents Chemother. 2025. PMID: 40323374 Free PMC article.
-
Imipenem-Relebactam and Meropenem-Vaborbactam: Two Novel Carbapenem-β-Lactamase Inhibitor Combinations.Drugs. 2018 Jan;78(1):65-98. doi: 10.1007/s40265-017-0851-9. Drugs. 2018. PMID: 29230684 Review.
-
A Systematic Review of the Pharmacokinetics and Pharmacodynamics of Novel Beta-Lactams and Beta-Lactam with Beta-Lactamase Inhibitor Combinations for the Treatment of Pneumonia Caused by Carbapenem-Resistant Gram-Negative Bacteria.Int J Antimicrob Agents. 2024 Sep;64(3):107266. doi: 10.1016/j.ijantimicag.2024.107266. Epub 2024 Jul 5. Int J Antimicrob Agents. 2024. PMID: 38971203
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI100560/AI/NIAID NIH HHS/United States
- PIP 11220200100191CO/Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET)
- R01 AI072219/AI/NIAID NIH HHS/United States
- R01 AI063517/AI/NIAID NIH HHS/United States
- PICT 2021-00771/MINCyT | Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT)
LinkOut - more resources
Full Text Sources
Miscellaneous

