Development and implementation of a core genome multilocus sequence typing scheme for Yersinia enterocolitica: a tool for surveillance and outbreak detection
- PMID: 38990041
- PMCID: PMC11325262
- DOI: 10.1128/jcm.00040-24
Development and implementation of a core genome multilocus sequence typing scheme for Yersinia enterocolitica: a tool for surveillance and outbreak detection
Abstract
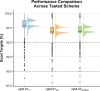
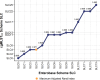
Yersinia enterocolitica (Y. enterocolitica) is the most frequent etiological agent of yersiniosis and has been responsible for several national outbreaks in Norway and elsewhere. A standardized high-resolution method, such as core genome Multilocus Sequence Typing (cgMLST), is needed for pathogen traceability at the national and international levels. In this study, we developed and implemented a cgMLST scheme for Y. enterocolitica. We designed a cgMLST scheme in SeqSphere + using high-quality genomes from different Y. enterocolitica biotype sublineages. The scheme was validated if more than 95% of targets were found across all tested Y. enterocolitica: 563 Norwegian genomes collected between 2012 and 2022 and 327 genomes from public data sets. We applied the scheme to known outbreaks to establish a threshold for identifying major complex types (CTs) based on the number of allelic differences. The final cgMLST scheme included 2,582 genes with a median of 97.9% (interquartile range 97.6%-98.8%) targets found across all tested genomes. Analysis of outbreaks identified all outbreak strains using single linkage clustering at four allelic differences. This threshold identified 311 unique CTs in Norway, of which CT18, CT12, and CT5 were identified as the most frequently associated with outbreaks. The cgMLST scheme showed a very good performance in typing Y. enterocolitica using diverse data sources and was able to identify outbreak clusters. We recommend the implementation of this scheme nationally and internationally to facilitate Y. enterocolitica surveillance and improve outbreak response in national and cross-border outbreaks.
Keywords: Yersinia enterocolitica; cgMLST; molecular typing; outbreak; yersiniosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Yersiniosis - annual epidemiological report for 2021. 2022. European Centre for Disease Prevention and Control. Available from: https://www.ecdc.europa.eu/en/publications-data/yersiniosis-annual-epide...
-
- Tack DM, Marder EP, Griffin PM, Cieslak PR, Dunn J, Hurd S, Scallan E, Lathrop S, Muse A, Ryan P, Smith K, Tobin-D’Angelo M, Vugia DJ, Holt KG, Wolpert BJ, Tauxe R, Geissler AL. 2019. Preliminary incidence and trends of infections with pathogens transmitted commonly through food-foodborne diseases active surveillance network, 10 U.S. sites, 2015-2018. MMWRMorb Mortal Wkly Rep 10:369–373. doi: 10.15585/mmwr.mm6816a2 - DOI - PMC - PubMed
-
- Guillier L, Fravalo P, Leclercq A, Thébault A, Kooh P, Cadavez V, Gonzales-Barron U. 2021. Risk factors for sporadic Yersinia enterocolitica infections: a systematic review and meta-analysis. Microb Risk Anal 17:100141. doi: 10.1016/j.mran.2020.100141 - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

