Substitutions in the nonactive site of the passenger domain on the activity of Haemophilus influenzae immunoglobulin A1 protease
- PMID: 38990045
- PMCID: PMC11320935
- DOI: 10.1128/iai.00193-24
Substitutions in the nonactive site of the passenger domain on the activity of Haemophilus influenzae immunoglobulin A1 protease
Abstract
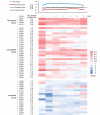
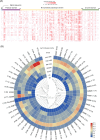
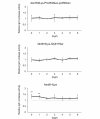
Immunoglobulin A1 (IgA1) protease is a critical virulence factor of Haemophilus influenzae that facilitates bacterial mucosal infection. This study investigates the effect of iga gene polymorphism on the enzymatic activity of H. influenzae IgA1 protease. The IgA1 protease activity was examined in the H. influenzae Rd KW20 strain and 51 isolates. Genetic variations in iga and deduced amino acid substitutions affecting IgA1 protease activity were assessed. Machine learning tools and functional complementation assays were used to analyze the effects of identified substitutions on the stability and activity of IgA1 protease, respectively. All 51 isolates exhibited similar iga expression levels. No igaB expression was detected. According to comparisons with the reference Rd KW20 strain, four substitutions in the protease domain, 26 in the nonprotease passenger domain, and two in the β-barrel domain were associated with the change in IgA1 protease activity. No substitutions in the catalytic site of IgA1 protease were observed. Logistic regression, receiver operating characteristic curves, Venn diagrams, and protein stability analyses revealed that the substitutions Asn352Lys, Pro353Ala, Lys356Asn, Gln916Lys, and Gly917Ser, which were located in the nonactive site of the passenger domain, were associated with decreases in IgA1 protease activity and stability, whereas Asn914Lys was associated with an increase in these events. Functional complementation assays revealed that the Asn914Lys substitution increased IgA1 protease activity in the Rd KW20 strain. This study identified substitutions in the nonactive site of the passenger domain that affect both the activity and stability of H. influenzae IgA1 protease.
Keywords: Haemophilus influenzae; immunoglobulin A1 protease; virulence factors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Characterization of igaB, a second immunoglobulin A1 protease gene in nontypeable Haemophilus influenzae.Infect Immun. 2006 Oct;74(10):5860-70. doi: 10.1128/IAI.00796-06. Infect Immun. 2006. PMID: 16988265 Free PMC article.
-
Nontypeable Haemophilus influenzae in carriage and disease: a difference in IgA1 protease activity levels.JAMA. 2002 Apr 3;287(13):1699-705. doi: 10.1001/jama.287.13.1699. JAMA. 2002. PMID: 11926894
-
Expression of IgA Proteases by Haemophilus influenzae in the Respiratory Tract of Adults With Chronic Obstructive Pulmonary Disease.J Infect Dis. 2015 Dec 1;212(11):1798-805. doi: 10.1093/infdis/jiv299. Epub 2015 May 20. J Infect Dis. 2015. PMID: 25995193 Free PMC article.
-
Antigenic and genetic heterogeneity among Haemophilus influenzae and Neisseria IgA1 proteases.Adv Exp Med Biol. 1995;371A:599-603. doi: 10.1007/978-1-4615-1941-6_126. Adv Exp Med Biol. 1995. PMID: 8525998 Review. No abstract available.
-
IgA1 protease.Int J Biochem Cell Biol. 2006;38(8):1244-8. doi: 10.1016/j.biocel.2005.10.005. Epub 2005 Nov 2. Int J Biochem Cell Biol. 2006. PMID: 16293440 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

