Variability in n-caprylate and n-caproate producing microbiomes in reactors with in-line product extraction
- PMID: 38990071
- PMCID: PMC11334527
- DOI: 10.1128/msystems.00416-24
Variability in n-caprylate and n-caproate producing microbiomes in reactors with in-line product extraction
Abstract
Medium-chain carboxylates (MCCs) are used in various industrial applications. These chemicals are typically extracted from palm oil, which is deemed not sustainable. Recent research has focused on microbial chain elongation using reactors to produce MCCs, such as n-caproate (C6) and n-caprylate (C8), from organic substrates such as wastes. Even though the production of n-caproate is relatively well-characterized, bacteria and metabolic pathways that are responsible for n-caprylate production are not. Here, three 5 L reactors with continuous membrane-based liquid-liquid extraction (i.e., pertraction) were fed ethanol and acetate and operated for an operating period of 234 days with different operating conditions. Metagenomic and metaproteomic analyses were employed. n-Caprylate production rates and reactor microbiomes differed between reactors even when operated similarly due to differences in H2 and O2 between the reactors. The complete reverse β-oxidation (RBOX) pathway was present and expressed by several bacterial species in the Clostridia class. Several Oscillibacter spp., including Oscillibacter valericigenes, were positively correlated with n-caprylate production rates, while Clostridium kluyveri was positively correlated with n-caproate production. Pseudoclavibacter caeni, which is a strictly aerobic bacterium, was abundant across all the operating periods, regardless of n-caprylate production rates. This study provides insight into microbiota that are associated with n-caprylate production in open-culture reactors and provides ideas for further work.IMPORTANCEMicrobial chain elongation pathways in open-culture biotechnology systems can be utilized to convert organic waste and industrial side streams into valuable industrial chemicals. Here, we investigated the microbiota and metabolic pathways that produce medium-chain carboxylates (MCCs), including n-caproate (C6) and n-caprylate (C8), in reactors with in-line product extraction. Although the reactors in this study were operated similarly, different microbial communities dominated and were responsible for chain elongation. We found that different microbiota were responsible for n-caproate or n-caprylate production, and this can inform engineers on how to operate the systems better. We also observed which changes in operating conditions steered the production toward and away from n-caprylate, but more work is necessary to ascertain a mechanistic understanding that could be predictive. This study provides pertinent research questions for future work.
Keywords: bacteria microcompartments; caproate; caprylate; chain elongation; hexanoate; hydrogen; medium-chain carboxylate; octanoate; oxygen.
Conflict of interest statement
L.T.A. has ownership in Capro-X, Inc., which is a start-up company that is commercializing a chain-elongating biotechnology production platform.
Figures
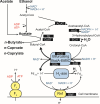
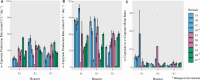
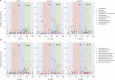


References
-
- Stamatopoulou P, Malkowski J, Conrado L, Brown K, Scarborough M. 2020. Fermentation of organic residues to beneficial chemicals: a review of medium-chain fatty acid production. Processes 8:1571. doi: 10.3390/pr8121571 - DOI
-
- Kucek LA, Spirito CM, Angenent LT. 2016. High n-caprylate productivities and specificities from dilute ethanol and acetate: chain elongation with microbiomes to upgrade products from syngas fermentation. Energy Environ Sci 9:3482–3494. doi: 10.1039/C6EE01487A - DOI
MeSH terms
Substances
Grants and funding
- STAR Fellowship/U.S. Environmental Protection Agency (EPA)
- Alexander von Humboldt Professorship/Alexander von Humboldt-Stiftung (AvH)
- EXC 2124 - 390838134/Deutsche Forschungsgemeinschaft (DFG)
- NNF21SA0072700/Novo Nordisk Fonden (NNF)
- W911NF-12-1-0555/DOD | USA | AFC | CCDC | Army Research Office (ARO)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
