Bacillus subtilis-derived peptides disrupt quorum sensing and biofilm assembly in multidrug-resistant Staphylococcus aureus
- PMID: 38990088
- PMCID: PMC11334493
- DOI: 10.1128/msystems.00712-24
Bacillus subtilis-derived peptides disrupt quorum sensing and biofilm assembly in multidrug-resistant Staphylococcus aureus
Abstract
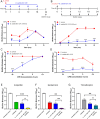
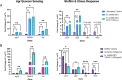
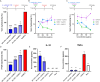
Multidrug-resistant Staphylococcus aureus is one of the most clinically important pathogens in the world, with infections leading to high rates of morbidity and mortality in both humans and animals. The ability of S. aureus to form biofilms protects cells from antibiotics and promotes the transfer of antibiotic resistance genes; therefore, new strategies aimed at inhibiting biofilm growth are urgently needed. Probiotic species, including Bacillus subtilis, are gaining interest as potential therapies against S. aureus for their ability to reduce S. aureus colonization and virulence. Here, we search for strains and microbially derived compounds with strong antibiofilm activity against multidrug-resistant S. aureus by isolating and screening Bacillus strains from a variety of agricultural environments. From a total of 1,123 environmental isolates, we identify a single strain B. subtilis 6D1, with a potent ability to inhibit biofilm growth, disassemble mature biofilm, and improve antibiotic sensitivity of S. aureus biofilms through an Agr quorum sensing interference mechanism. Biochemical and molecular networking analysis of an active organic fraction revealed multiple surfactin isoforms, and an uncharacterized peptide was driving this antibiofilm activity. Compared with commercial high-performance liquid chromatography grade surfactin obtained from B. subtilis, we show these B. subtilis 6D1 peptides are significantly better at inhibiting biofilm formation in all four S. aureus Agr backgrounds and preventing S. aureus-induced cytotoxicity when applied to HT29 human intestinal cells. Our study illustrates the potential of exploring microbial strain diversity to discover novel antibiofilm agents that may help combat multidrug-resistant S. aureus infections and enhance antibiotic efficacy in clinical and veterinary settings.
Importance: The formation of biofilms by multidrug-resistant bacterial pathogens, such as Staphylococcus aureus, increases these microorganisms' virulence and decreases the efficacy of common antibiotic regimens. Probiotics possess a variety of strain-specific strategies to reduce biofilm formation in competing organisms; however, the mechanisms and compounds responsible for these phenomena often go uncharacterized. In this study, we identified a mixture of small probiotic-derived peptides capable of Agr quorum sensing interference as one of the mechanisms driving antibiofilm activity against S. aureus. This collection of peptides also improved antibiotic killing and protected human gut epithelial cells from S. aureus-induced toxicity by stimulating an adaptive cytokine response. We conclude that purposeful strain screening and selection efforts can be used to identify unique probiotic strains that possess specially desired mechanisms of action. This information can be used to further improve our understanding of the ways in which probiotic and probiotic-derived compounds can be applied to prevent bacterial infections or improve bacterial sensitivity to antibiotics in clinical and agricultural settings.
Keywords: Bacillus subtilis; Staphylococcus aureus; antibiotic resistance; biofilm; peptide; postbiotic; probiotic; quorum sensing interference.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Disarming Staphylococcus aureus: Review of Strategies Combating This Resilient Pathogen by Targeting Its Virulence.Pathogens. 2025 Apr 15;14(4):386. doi: 10.3390/pathogens14040386. Pathogens. 2025. PMID: 40333163 Free PMC article. Review.
-
Unveilling genetic profiles and correlations of biofilm-associated genes, quorum sensing, and antibiotic resistance in Staphylococcus aureus isolated from a Malaysian Teaching Hospital.Eur J Med Res. 2024 Apr 22;29(1):246. doi: 10.1186/s40001-024-01831-6. Eur J Med Res. 2024. PMID: 38649897 Free PMC article.
-
Dispersal of pathogen-associated multispecies biofilm by novel probiotic Bacillus subtilis in a contact-dependent manner.J Appl Microbiol. 2022 Oct;133(4):2501-2515. doi: 10.1111/jam.15721. Epub 2022 Aug 6. J Appl Microbiol. 2022. PMID: 35858688
-
Baohuoside I inhibits virulence of multidrug-resistant Staphylococcus aureus by targeting the transcription Staphylococcus accessory regulator factor SarZ.Phytomedicine. 2024 Jul 25;130:155590. doi: 10.1016/j.phymed.2024.155590. Epub 2024 Apr 2. Phytomedicine. 2024. PMID: 38810547
-
Focused review on dual inhibition of quorum sensing and efflux pumps: A potential way to combat multi drug resistant Staphylococcus aureus infections.Int J Biol Macromol. 2021 Nov 1;190:33-43. doi: 10.1016/j.ijbiomac.2021.08.199. Epub 2021 Sep 1. Int J Biol Macromol. 2021. PMID: 34480904 Review.
Cited by
-
Disarming Staphylococcus aureus: Review of Strategies Combating This Resilient Pathogen by Targeting Its Virulence.Pathogens. 2025 Apr 15;14(4):386. doi: 10.3390/pathogens14040386. Pathogens. 2025. PMID: 40333163 Free PMC article. Review.
-
Population and pan-genomic analyses of Staphylococcus pseudintermedius identify geographic distinctions in accessory gene content and novel loci associated with AMR.Appl Environ Microbiol. 2025 May 21;91(5):e0001025. doi: 10.1128/aem.00010-25. Epub 2025 Apr 24. Appl Environ Microbiol. 2025. PMID: 40272117 Free PMC article.
-
Antibacterial and anticancer activities of rhizobacterial lipopeptides against carcinogenic bacteria.Antonie Van Leeuwenhoek. 2025 Jul 21;118(9):118. doi: 10.1007/s10482-025-02131-7. Antonie Van Leeuwenhoek. 2025. PMID: 40691732
-
Discovery of a Potent Antimicrobial Peptide Through Rational Design: A New Frontier in Pathogen Control.Biomolecules. 2025 Jul 11;15(7):989. doi: 10.3390/biom15070989. Biomolecules. 2025. PMID: 40723861 Free PMC article.
-
Genome Mining Reveals a Sactipeptide Biosynthetic Cluster in Staphylococcus pseudintermedius.Vet Sci. 2025 Jul 2;12(7):635. doi: 10.3390/vetsci12070635. Vet Sci. 2025. PMID: 40711295 Free PMC article.
References
-
- Centers for Disease Control and Prevention (U.S) . 2019. Antibiotic resistance threats in the United States, 2019. Centers for Disease Control and Prevention (U.S.)
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
