Integrative gene expression analysis and animal model reveal immune- and autophagy-related biomarkers in osteomyelitis
- PMID: 38990187
- PMCID: PMC11238574
- DOI: 10.1002/iid3.1339
Integrative gene expression analysis and animal model reveal immune- and autophagy-related biomarkers in osteomyelitis
Abstract
Background: Osteomyelitis (OM) is recognized as a significant challenge in orthopedics due to its complex immune and inflammatory responses. The prognosis heavily depends on timely diagnosis, accurate classification, and assessment of severity. Thus, the identification of diagnostic and classification-related genes from an immunological standpoint is crucial for the early detection and tailored treatment of OM.
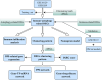
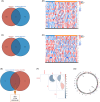
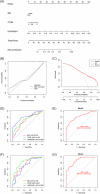
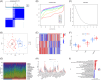
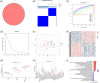
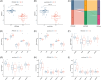
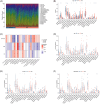
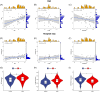
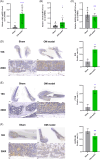
Methods: Transcriptomic data for OM was sourced from the Gene Expression Omnibus (GEO) database, leading to the identification of autophagy- and immune-related differentially expressed genes (AIR-DEGs) through differential expression analysis. Diagnostic and classification models were subsequently developed. The CIBERSORT algorithm was utilized to examine immune cell infiltration in OM, and the relationship between OM clusters and various immune cells was explored. Key AIR-DEGs were further validated through the creation of OM animal models.
Results: Analysis of the transcriptomic data revealed three AIR-DEGs that played a significant role in immune responses and pathways. Nomogram and receiver operating characteristic curve analyses were performed, demonstrating excellent diagnostic capability for differentiating between OM patients and healthy individuals, with an area under the curve of 0.814. An unsupervised clustering analysis discerned two unique patterns of autophagy- and immune-related genes, as well as gene patterns. Further exploration into immune infiltration exhibited notable variances across different subtypes, especially between OM cluster 1 and gene cluster A, highlighting their potential role in mitigating inflammatory responses by regulating immune activities. Moreover, the mRNA and protein expression levels of three AIR-DEGs in the animal model were aligned with those in the training and validation data sets.
Conclusions: From an immunological perspective, a diagnostic model was successfully developed, and two distinct clustering patterns were identified. These contributions offer a significant resource for the early detection and personalized immunotherapy of patients with OM.
Keywords: autophagy; clustering pattern; diagnosis; immune; immune infiltration; osteomyelitis.
© 2024 The Author(s). Immunity, Inflammation and Disease published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Lew DP, Waldvogel FA. Osteomyelitis. Lancet. 2004;364(9431):369‐379. - PubMed
-
- Momodu II, Savaliya V. Osteomyelitis. StatPearls. StatPearls Publishing Copyright © 2022, StatPearls Publishing LLC.; 2022.
-
- Hofstee MI, Muthukrishnan G, Atkins GJ, et al. Current concepts of osteomyelitis. Am J Pathol. 2020;190(6):1151‐1163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

