Mmp14-dependent remodeling of the pericellular-dermal collagen interface governs fibroblast survival
- PMID: 38990714
- PMCID: PMC11244150
- DOI: 10.1083/jcb.202312091
Mmp14-dependent remodeling of the pericellular-dermal collagen interface governs fibroblast survival
Abstract
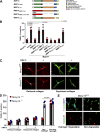
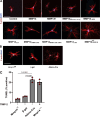
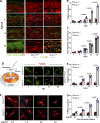
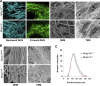
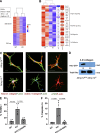
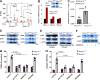
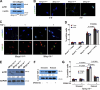
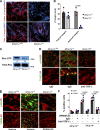
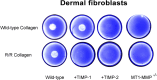
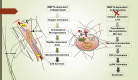
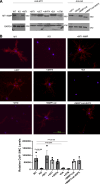
Dermal fibroblasts deposit type I collagen, the dominant extracellular matrix molecule found in skin, during early postnatal development. Coincident with this biosynthetic program, fibroblasts proteolytically remodel pericellular collagen fibrils by mobilizing the membrane-anchored matrix metalloproteinase, Mmp14. Unexpectedly, dermal fibroblasts in Mmp14-/- mice commit to a large-scale apoptotic program that leaves skin tissues replete with dying cells. A requirement for Mmp14 in dermal fibroblast survival is recapitulated in vitro when cells are embedded within, but not cultured atop, three-dimensional hydrogels of crosslinked type I collagen. In the absence of Mmp14-dependent pericellular proteolysis, dermal fibroblasts fail to trigger β1 integrin activation and instead actuate a TGF-β1/phospho-JNK stress response that leads to apoptotic cell death in vitro as well as in vivo. Taken together, these studies identify Mmp14 as a requisite cell survival factor that maintains dermal fibroblast viability in postnatal dermal tissues.
© 2024 Sabeh et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures
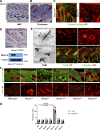
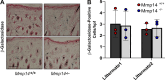
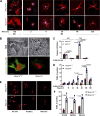
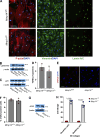
References
-
- Alabi, A., Xia X.-D., Gu H.-M., Wang F., Deng S.-J., Yang N., Adijiang A., Douglas D.N., Kneteman N.M., Xue Y., et al. . 2021. Membrane type 1 matrix metalloproteinase promotes LDL receptor shedding and accelerates the development of atherosclerosis. Nat. Commun. 12:1889. 10.1038/s41467-021-22167-3 - DOI - PMC - PubMed
-
- Andrews, S. 2010. FastQC: A Quality Control Tool for High Throughput Sequence Data.
-
- Birukawa, N.K., Murase K., Sato Y., Kosaka A., Yoneda A., Nishita H., Fujita R., Nishimura M., Ninomiya T., Kajiwara K., et al. . 2014. Activated hepatic stellate cells are dependent on self-collagen, cleaved by membrane type 1 matrix metalloproteinase for their growth. J. Biol. Chem. 289:20209–20221. 10.1074/jbc.M113.544494 - DOI - PMC - PubMed
-
- Bouffard, N.A., Cutroneo K.R., Badger G.J., White S.L., Buttolph T.R., Ehrlich H.P., Stevens-Tuttle D., and Langevin H.M.. 2008. Tissue stretch decreases soluble TGF-beta1 and type-1 procollagen in mouse subcutaneous connective tissue: Evidence from ex vivo and in vivo models. J. Cell. Physiol. 214:389–395. 10.1002/jcp.21209 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

