Nicotinamide riboside Induced Energy Stress and Metabolic Reprogramming in BEAS-2B Cells
- PMID: 38990804
- PMCID: PMC11337214
- DOI: 10.1021/acs.chemrestox.3c00312
Nicotinamide riboside Induced Energy Stress and Metabolic Reprogramming in BEAS-2B Cells
Abstract
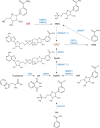
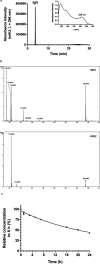
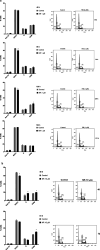
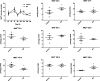
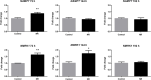
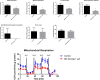
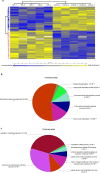
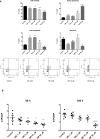
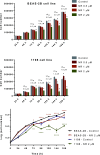
Nicotinamide riboside (NR), a NAD+ precursor, has received attention due to several health benefits it has induced in experimental models. Studies in cultured cells, animals, and humans consistently show increased NAD+ availability after NR supplementation, which is considered the only mode of NR action that leads to health benefits. In the present study, we show that a persistently low NR concentration (1 μM) in the growth medium of BEAS-2B human cells, grown in a monolayer, induces energy stress, which precedes a cellular NAD+ increase after 192 h. NR concentrations greater than 1 μM under the specified conditions were cytotoxic in the 2D cell culture model, while all concentrations tested in the 3D cell culture model (BEAS-2B cell spheroids exposed to 1, 5, 10, and 50 μM NR) induced apoptosis. Shotgun proteomics revealed that NR modulated the abundance of proteins, agreeing with the observed effects on cellular energy metabolism and cell growth or survival. Energy stress may activate pathways that lead to health benefits such as cancer prevention. Accordingly, the premalignant 1198 cell line was more sensitive to NR cytotoxicity than the phenotypically normal parent BEAS-2B cell line. The role of a mild energy stress induced by low concentrations of NR in its beneficial effects deserves further investigation. On the other hand, strategies to increase the bioavailability of NR require attention to toxic effects that may arise.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




Similar articles
-
Nicotinamide Riboside Enhances In Vitro Beta-adrenergic Brown Adipose Tissue Activity in Humans.J Clin Endocrinol Metab. 2021 Apr 23;106(5):1437-1447. doi: 10.1210/clinem/dgaa960. J Clin Endocrinol Metab. 2021. PMID: 33524145 Clinical Trial.
-
Nicotinamide riboside, an NAD+ precursor, attenuates the development of liver fibrosis in a diet-induced mouse model of liver fibrosis.Biochim Biophys Acta Mol Basis Dis. 2019 Sep 1;1865(9):2451-2463. doi: 10.1016/j.bbadis.2019.06.009. Epub 2019 Jun 11. Biochim Biophys Acta Mol Basis Dis. 2019. PMID: 31195117 Free PMC article.
-
Targeting sirtuin activity with nicotinamide riboside reduces neuroinflammation in a GWI mouse model.Neurotoxicology. 2020 Jul;79:84-94. doi: 10.1016/j.neuro.2020.04.006. Epub 2020 Apr 25. Neurotoxicology. 2020. PMID: 32343995
-
Balancing NAD+ deficits with nicotinamide riboside: therapeutic possibilities and limitations.Cell Mol Life Sci. 2022 Aug 2;79(8):463. doi: 10.1007/s00018-022-04499-5. Cell Mol Life Sci. 2022. PMID: 35918544 Free PMC article. Review.
-
Nicotinamide Riboside-The Current State of Research and Therapeutic Uses.Nutrients. 2020 May 31;12(6):1616. doi: 10.3390/nu12061616. Nutrients. 2020. PMID: 32486488 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

