An Orphan Gene Enhances Male Reproductive Success in Plutella xylostella
- PMID: 38990889
- PMCID: PMC11290247
- DOI: 10.1093/molbev/msae142
An Orphan Gene Enhances Male Reproductive Success in Plutella xylostella
Abstract
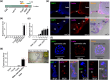
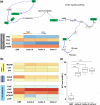
Plutella xylostella exhibits exceptional reproduction ability, yet the genetic basis underlying the high reproductive capacity remains unknown. Here, we demonstrate that an orphan gene, lushu, which encodes a sperm protein, plays a crucial role in male reproductive success. Lushu is located on the Z chromosome and is prevalent across different P. xylostella populations worldwide. We subsequently generated lushu mutants using transgenic CRISPR/Cas9 system. Knockout of Lushu results in reduced male mating efficiency and accelerated death in adult males. Furthermore, our findings highlight that the deficiency of lushu reduced the transfer of sperms from males to females, potentially resulting in hindered sperm competition. Additionally, the knockout of Lushu results in disrupted gene expression in energy-related pathways and elevated insulin levels in adult males. Our findings reveal that male reproductive performance has evolved through the birth of a newly evolved, lineage-specific gene with enormous potentiality in fecundity success. These insights hold valuable implications for identifying the target for genetic control, particularly in relation to species-specific traits that are pivotal in determining high levels of fecundity.
Keywords: insulin; male reproductive fitness; orphan gene; sperm competition; sperm protein.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
Conflict of Interest None declared.
Figures



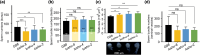

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

