Nanopore tweezers show fractional-nucleotide translocation in sequence-dependent pausing by RNA polymerase
- PMID: 38990947
- PMCID: PMC11260103
- DOI: 10.1073/pnas.2321017121
Nanopore tweezers show fractional-nucleotide translocation in sequence-dependent pausing by RNA polymerase
Abstract
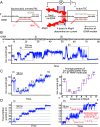
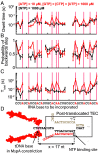
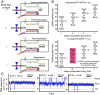
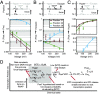
RNA polymerases (RNAPs) carry out the first step in the central dogma of molecular biology by transcribing DNA into RNA. Despite their importance, much about how RNAPs work remains unclear, in part because the small (3.4 Angstrom) and fast (~40 ms/nt) steps during transcription were difficult to resolve. Here, we used high-resolution nanopore tweezers to observe the motion of single Escherichia coli RNAP molecules as it transcribes DNA ~1,000 times improved temporal resolution, resolving single-nucleotide and fractional-nucleotide steps of individual RNAPs at saturating nucleoside triphosphate concentrations. We analyzed RNAP during processive transcription elongation and sequence-dependent pausing at the yrbL elemental pause sequence. Each time RNAP encounters the yrbL elemental pause sequence, it rapidly interconverts between five translocational states, residing predominantly in a half-translocated state. The kinetics and force-dependence of this half-translocated state indicate it is a functional intermediate between pre- and post-translocated states. Using structural and kinetics data, we show that, in the half-translocated and post-translocated states, sequence-specific protein-DNA interaction occurs between RNAP and a guanine base at the downstream end of the transcription bubble (core recognition element). Kinetic data show that this interaction stabilizes the half-translocated and post-translocated states relative to the pre-translocated state. We develop a kinetic model for RNAP at the yrbL pause and discuss this in the context of key structural features.
Keywords: enzymology; nanopore tweezers; transcription; transcription elongation; transcription pausing.
Conflict of interest statement
Competing interests statement:J.M.C., A.H.L., I.M.D., H.B., and J.H.G. hold US patent 10,359,395 on the Nanopore Tweezers technology.
Figures



Similar articles
-
RNA Polymerase Accommodates a Pause RNA Hairpin by Global Conformational Rearrangements that Prolong Pausing.Mol Cell. 2018 Mar 1;69(5):802-815.e5. doi: 10.1016/j.molcel.2018.01.018. Mol Cell. 2018. PMID: 29499135 Free PMC article.
-
Applied force provides insight into transcriptional pausing and its modulation by transcription factor NusA.Mol Cell. 2011 Nov 18;44(4):635-46. doi: 10.1016/j.molcel.2011.09.018. Mol Cell. 2011. PMID: 22099310 Free PMC article.
-
Visualizing translocation dynamics and nascent transcript errors in paused RNA polymerases in vivo.Genome Biol. 2015 May 15;16(1):98. doi: 10.1186/s13059-015-0666-5. Genome Biol. 2015. PMID: 25976475 Free PMC article.
-
Transcription factor dynamics.Microbiology (Reading). 2008 Jul;154(Pt 7):1837-1844. doi: 10.1099/mic.0.2008/018549-0. Microbiology (Reading). 2008. PMID: 18599813 Review.
-
Transcription elongation.Transcription. 2017 May 27;8(3):150-161. doi: 10.1080/21541264.2017.1289294. Epub 2017 Feb 8. Transcription. 2017. PMID: 28301288 Free PMC article. Review.
Cited by
-
RNA Polymerase II Activity Control of Gene Expression and Involvement in Disease.J Mol Biol. 2025 Jan 1;437(1):168770. doi: 10.1016/j.jmb.2024.168770. Epub 2024 Aug 28. J Mol Biol. 2025. PMID: 39214283 Free PMC article. Review.
-
The potential of fluorogenicity for single molecule FRET and DyeCycling.QRB Discov. 2024 Dec 3;5:e8. doi: 10.1017/qrd.2024.11. eCollection 2024. QRB Discov. 2024. PMID: 39687231 Free PMC article. Review.
References
-
- Erie D. A., Yager T. D., von Hippel P. H., The single-nucleotide addition cycle in transcription: A biophysical and biochemical perspective. Annu. Rev. Biophys. Biomol. Struct. 21, 379–415 (1992). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

