Germline BARD1 variants predispose to mesothelioma by impairing DNA repair and calcium signaling
- PMID: 38990952
- PMCID: PMC11260134
- DOI: 10.1073/pnas.2405231121
Germline BARD1 variants predispose to mesothelioma by impairing DNA repair and calcium signaling
Abstract
We report that ~1.8% of all mesothelioma patients and 4.9% of those younger than 55, carry rare germline variants of the BRCA1 associated RING domain 1 (BARD1) gene that were predicted to be damaging by computational analyses. We conducted functional assays, essential for accurate interpretation of missense variants, in primary fibroblasts that we established in tissue culture from a patient carrying the heterozygous BARD1V523A mutation. We found that these cells had genomic instability, reduced DNA repair, and impaired apoptosis. Investigating the underlying signaling pathways, we found that BARD1 forms a trimeric protein complex with p53 and SERCA2 that regulates calcium signaling and apoptosis. We validated these findings in BARD1-silenced primary human mesothelial cells exposed to asbestos. Our study elucidated mechanisms of BARD1 activity and revealed that heterozygous germline BARD1 mutations favor the development of mesothelioma and increase the susceptibility to asbestos carcinogenesis. These mesotheliomas are significantly less aggressive compared to mesotheliomas in asbestos workers.
Keywords: cancer prevention; carcinogenesis; gene × environment; genetics; mesothelioma.
Conflict of interest statement
Competing interests statement:M.C. has a patent issued for “Methods for Diagnosing a Predisposition to Develop Cancer.” M.C. and H.Y. have a patent issued for “Using Anti-HMGB1 Monoclonal Antibody or other HMGB1 Antibodies as a Novel Mesothelioma Therapeutic Strategy”, and a patent issued for “HMGB1 As a Biomarker for Asbestos Exposure and Mesothelioma Early Detection.” M.C. is a board-certified pathologist who provides consultation for pleural pathology, including medical-legal.
Figures

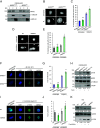
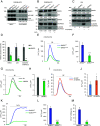
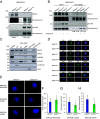
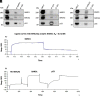


References
-
- Zhao J., The global trends in incidence, death, burden and risk factors of early-onset cancer from 1990 to 2019. BMJ Oncol. 2, 12 (2023).
-
- Carbone M., Yang H., Pass H. I., Taioli E., Did the Ban on asbestos reduce the incidence of mesothelioma? J. Thorac. Oncol. 18, 694–697 (2023). - PubMed
-
- Roushdy-Hammady I., Siegel J., Emri S., Testa J. R., Carbone M., Genetic-susceptibility factor and malignant mesothelioma in the Cappadocian region of Turkey. Lancet 357, 444–445 (2001). - PubMed
-
- Carbone M., et al. , A mesothelioma epidemic in Cappadocia: Scientific developments and unexpected social outcomes. Nat. Rev. Cancer 7, 147–154 (2007). - PubMed
MeSH terms
Substances
Grants and funding
- R01 CA198138/CA/NCI NIH HHS/United States
- S10 OD028515/OD/NIH HHS/United States
- 853057/ERC_/European Research Council/International
- IG-19803/Italian Association for Cancer Research
- PRIN2017E5L5P3 t/AROSE, Progetti di Rilevante Interesse Nazionale
- C-1792/Welch Foundation (The Welch Foundation)
- GR-2013-02356747/Italian Ministry of Health
- R01 ES030948/ES/NIEHS NIH HHS/United States
- 5U01CA214195-04/the Early Detection Research Network NCI
- 1R01ES030948-01/HHS | NIH | National Institute of Environmental Health Sciences (NIEHS)
- IG-23670/Italian Association for Cancer Research
- 1R01CA237235-01A1/HHS | NIH | National Cancer Institute (NCI)
- 1R01CA198138/US Department of Defence
- R01 CA237235/CA/NCI NIH HHS/United States
- PHY-2019745/NSF
- U01 CA214195/CA/NCI NIH HHS/United States
- PRIN20177E9EPY/AROSE, Progetti di Rilevante Interesse Nazionale
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

