Structural and genetic diversity in the secreted mucins MUC5AC and MUC5B
- PMID: 38991590
- PMCID: PMC11344006
- DOI: 10.1016/j.ajhg.2024.06.007
Structural and genetic diversity in the secreted mucins MUC5AC and MUC5B
Abstract
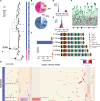
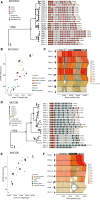
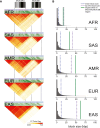
The secreted mucins MUC5AC and MUC5B are large glycoproteins that play critical defensive roles in pathogen entrapment and mucociliary clearance. Their respective genes contain polymorphic and degenerate protein-coding variable number tandem repeats (VNTRs) that make the loci difficult to investigate with short reads. We characterize the structural diversity of MUC5AC and MUC5B by long-read sequencing and assembly of 206 human and 20 nonhuman primate (NHP) haplotypes. We find that human MUC5B is largely invariant (5,761-5,762 amino acids [aa]); however, seven haplotypes have expanded VNTRs (6,291-7,019 aa). In contrast, 30 allelic variants of MUC5AC encode 16 distinct proteins (5,249-6,325 aa) with cysteine-rich domain and VNTR copy-number variation. We group MUC5AC alleles into three phylogenetic clades: H1 (46%, ∼5,654 aa), H2 (33%, ∼5,742 aa), and H3 (7%, ∼6,325 aa). The two most common human MUC5AC variants are smaller than NHP gene models, suggesting a reduction in protein length during recent human evolution. Linkage disequilibrium and Tajima's D analyses reveal that East Asians carry exceptionally large blocks with an excess of rare variation (p < 0.05) at MUC5AC. To validate this result, we use Locityper for genotyping MUC5AC haplogroups in 2,600 unrelated samples from the 1000 Genomes Project. We observe a signature of positive selection in H1 among East Asians and a depletion of the likely ancestral haplogroup (H3). In Europeans, H3 alleles show an excess of common variation and deviate from Hardy-Weinberg equilibrium (p < 0.05), consistent with heterozygote advantage and balancing selection. This study provides a generalizable strategy to characterize complex protein-coding VNTRs for improved disease associations.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests E.E.E. is a scientific advisory board (SAB) member of Variant Bio, Inc.
Figures



Update of
-
Structural and genetic diversity in the secreted mucins, MUC5AC and MUC5B.bioRxiv [Preprint]. 2024 Mar 20:2024.03.18.585560. doi: 10.1101/2024.03.18.585560. bioRxiv. 2024. Update in: Am J Hum Genet. 2024 Aug 8;111(8):1700-1716. doi: 10.1016/j.ajhg.2024.06.007. PMID: 38562829 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

