Dual-localized PPTC7 limits mitophagy through proximal and dynamic interactions with BNIP3 and NIX
- PMID: 38991726
- PMCID: PMC11239977
- DOI: 10.26508/lsa.202402765
Dual-localized PPTC7 limits mitophagy through proximal and dynamic interactions with BNIP3 and NIX
Abstract
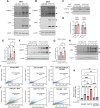
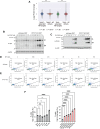
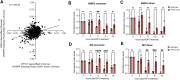
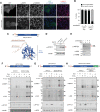
PPTC7 is a mitochondrial-localized phosphatase that suppresses BNIP3- and NIX-mediated mitophagy, but the mechanisms underlying this regulation remain ill-defined. Here, we demonstrate that loss of PPTC7 upregulates BNIP3 and NIX post-transcriptionally and independent of HIF-1α stabilization. Loss of PPTC7 prolongs the half-life of BNIP3 and NIX while blunting their accumulation in response to proteasomal inhibition, suggesting that PPTC7 promotes the ubiquitin-mediated turnover of BNIP3 and NIX. Consistently, overexpression of PPTC7 limits the accumulation of BNIP3 and NIX protein levels, which requires an intact catalytic motif but is surprisingly independent of its targeting to mitochondria. Consistently, we find that PPTC7 is dual-localized to the outer mitochondrial membrane and the matrix. Importantly, anchoring PPTC7 to the outer mitochondrial membrane is sufficient to blunt BNIP3 and NIX accumulation, and proximity labeling and fluorescence co-localization experiments demonstrate that PPTC7 dynamically associates with BNIP3 and NIX within the native cellular environment. Collectively, these data reveal that a fraction of PPTC7 localizes to the outer mitochondrial membrane to promote the proteasomal turnover of BNIP3 and NIX, limiting basal mitophagy.
© 2024 Wei et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Update of
-
PPTC7 limits mitophagy through proximal and dynamic interactions with BNIP3 and NIX.bioRxiv [Preprint]. 2024 Jan 25:2024.01.24.576953. doi: 10.1101/2024.01.24.576953. bioRxiv. 2024. Update in: Life Sci Alliance. 2024 Jul 11;7(9):e202402765. doi: 10.26508/lsa.202402765. PMID: 38328188 Free PMC article. Updated. Preprint.
Similar articles
-
Coordinating BNIP3/NIX-mediated mitophagy in space and time.Biochem Soc Trans. 2024 Oct 30;52(5):1969-1979. doi: 10.1042/BST20221364. Biochem Soc Trans. 2024. PMID: 39377319 Free PMC article. Review.
-
PPTC7 limits mitophagy through proximal and dynamic interactions with BNIP3 and NIX.bioRxiv [Preprint]. 2024 Jan 25:2024.01.24.576953. doi: 10.1101/2024.01.24.576953. bioRxiv. 2024. Update in: Life Sci Alliance. 2024 Jul 11;7(9):e202402765. doi: 10.26508/lsa.202402765. PMID: 38328188 Free PMC article. Updated. Preprint.
-
PPTC7 antagonizes mitophagy by promoting BNIP3 and NIX degradation via SCFFBXL4.EMBO Rep. 2024 Aug;25(8):3324-3347. doi: 10.1038/s44319-024-00181-y. Epub 2024 Jul 11. EMBO Rep. 2024. PMID: 38992176 Free PMC article.
-
A mitophagy sensor PPTC7 controls BNIP3 and NIX degradation to regulate mitochondrial mass.Mol Cell. 2024 Jan 18;84(2):327-344.e9. doi: 10.1016/j.molcel.2023.11.038. Epub 2023 Dec 26. Mol Cell. 2024. PMID: 38151018
-
Mitochondrial autophagy: Origins, significance, and role of BNIP3 and NIX.Biochim Biophys Acta. 2015 Oct;1853(10 Pt B):2775-83. doi: 10.1016/j.bbamcr.2015.02.022. Epub 2015 Mar 6. Biochim Biophys Acta. 2015. PMID: 25753537 Review.
Cited by
-
Calcium-mediated regulation of mitophagy: implications in neurodegenerative diseases.NPJ Metab Health Dis. 2025;3(1):4. doi: 10.1038/s44324-025-00049-2. Epub 2025 Feb 3. NPJ Metab Health Dis. 2025. PMID: 39911695 Free PMC article. Review.
-
Coordinating BNIP3/NIX-mediated mitophagy in space and time.Biochem Soc Trans. 2024 Oct 30;52(5):1969-1979. doi: 10.1042/BST20221364. Biochem Soc Trans. 2024. PMID: 39377319 Free PMC article. Review.
-
Legionella effector LpPIP recruits protein phosphatase 1 to the mitochondria to induce dephosphorylation of outer membrane proteins.PLoS Biol. 2025 Jul 23;23(7):e3003261. doi: 10.1371/journal.pbio.3003261. eCollection 2025 Jul. PLoS Biol. 2025. PMID: 40700448 Free PMC article.
-
Reconstitution of BNIP3/NIX-mitophagy initiation reveals hierarchical flexibility of the autophagy machinery.Nat Cell Biol. 2025 Aug;27(8):1272-1287. doi: 10.1038/s41556-025-01712-y. Epub 2025 Jul 25. Nat Cell Biol. 2025. PMID: 40715440 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
