Natural variation of TBR confers plant zinc toxicity tolerance through root cell wall pectin methylesterification
- PMID: 38992052
- PMCID: PMC11239920
- DOI: 10.1038/s41467-024-50106-5
Natural variation of TBR confers plant zinc toxicity tolerance through root cell wall pectin methylesterification
Abstract
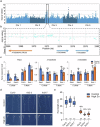
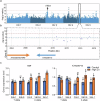
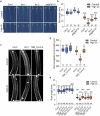
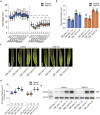
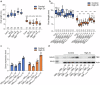
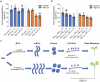
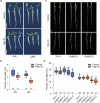
Zinc (Zn) is an essential micronutrient but can be cytotoxic when present in excess. Plants have evolved mechanisms to tolerate Zn toxicity. To identify genetic loci responsible for natural variation of plant tolerance to Zn toxicity, we conduct genome-wide association studies for root growth responses to high Zn and identify 21 significant associated loci. Among these loci, we identify Trichome Birefringence (TBR) allelic variation determining root growth variation in high Zn conditions. Natural alleles of TBR determine TBR transcript and protein levels which affect pectin methylesterification in root cell walls. Together with previously published data showing that pectin methylesterification increase goes along with decreased Zn binding to cell walls in TBR mutants, our findings lead to a model in which TBR allelic variation enables Zn tolerance through modulating root cell wall pectin methylesterification. The role of TBR in Zn tolerance is conserved across dicot and monocot plant species.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
LEUNIG_HOMOLOG transcriptional co-repressor mediates aluminium sensitivity through PECTIN METHYLESTERASE46-modulated root cell wall pectin methylesterification in Arabidopsis.Plant J. 2017 May;90(3):491-504. doi: 10.1111/tpj.13506. Epub 2017 Mar 25. Plant J. 2017. PMID: 28181322
-
A mutation in the Arabidopsis thaliana cell wall biosynthesis gene pectin methylesterase 3 as well as its aberrant expression cause hypersensitivity specifically to Zn.Plant J. 2013 Oct;76(1):151-64. doi: 10.1111/tpj.12279. Epub 2013 Aug 5. Plant J. 2013. PMID: 23826687
-
Pectin Methylesterification Impacts the Relationship between Photosynthesis and Plant Growth.Plant Physiol. 2016 Jun;171(2):833-48. doi: 10.1104/pp.16.00173. Epub 2016 Apr 4. Plant Physiol. 2016. PMID: 27208234 Free PMC article.
-
The zinc homeostasis network of land plants.Biochim Biophys Acta. 2012 Sep;1823(9):1553-67. doi: 10.1016/j.bbamcr.2012.05.016. Epub 2012 May 22. Biochim Biophys Acta. 2012. PMID: 22626733 Review.
-
Tuning of pectin methylesterification: consequences for cell wall biomechanics and development.Planta. 2015 Oct;242(4):791-811. doi: 10.1007/s00425-015-2358-5. Epub 2015 Jul 14. Planta. 2015. PMID: 26168980 Review.
Cited by
-
Changing paradigms for the micronutrient zinc, a known protein cofactor, as a signal relaying also cellular redox state.Quant Plant Biol. 2025 Apr 2;6:e7. doi: 10.1017/qpb.2025.4. eCollection 2025. Quant Plant Biol. 2025. PMID: 40297241 Free PMC article. Review.
References
-
- Nagajyoti PC, Lee KD, Sreekanth TVM. Heavy metals, occurrence and toxicity for plants: a review. Environ. chem. Lett. 2010;8:199–216. doi: 10.1007/s10311-010-0297-8. - DOI
-
- Marschner, P. Mineral Nutrition of Higher Plants 3rd edn, (London: Academic Press, 2012).
-
- Chaney, R. L. Zinc phytotoxicity. In: Robson A. D., ed. Zinc in soil and plants. Dordrecht, the Netherlands: Kluwer Academic Publishers, 135–150 (1993).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

