Epigenetically associated IGF2BP3 upregulation promotes cell proliferation by regulating E2F1 expression in hepatocellular carcinoma
- PMID: 38992083
- PMCID: PMC11239653
- DOI: 10.1038/s41598-024-67021-w
Epigenetically associated IGF2BP3 upregulation promotes cell proliferation by regulating E2F1 expression in hepatocellular carcinoma
Abstract
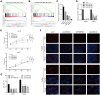
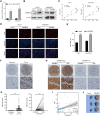
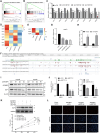
RNA-binding proteins (RBPs) are a class of proteins that primarily function by interacting with different types of RNAs and play a critical role in regulating the transcription and translation of cancer-related genes. However, their role in the progression of hepatocellular carcinoma (HCC) remains unclear. In this study, we analyzed RNA sequencing data and the corresponding clinical information of patients with HCC to screen for prognostic RBPs. Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3) was identified as an independent prognostic factor for liver cancer. It is upregulated in HCC and is associated with a poor prognosis. Elevated IGF2BP3 expression was validated via immunohistochemical analysis using a tissue microarray of patients with HCC. IGF2BP3 knockdown inhibited the proliferation of Hep3B and HepG2 cells, whereas IGF2BP3 overexpression promoted the expansion of HuH-7 and MHCC97H cells. Mechanistically, IGF2BP3 modulates cell proliferation by regulating E2F1 expression. DNA hypomethylation of the IGF2BP3 gene may increase the expression of IGF2BP3, thereby enhancing cell proliferation in HCC. Therefore, IGF2BP3 may act as a novel prognostic biomarker and a potential therapeutic target for HCC.
Keywords: E2F1; HCC; Methylation; Prognosis; RNA-binding protein.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

