A near complete genome assembly of the East Friesian sheep genome
- PMID: 38992134
- PMCID: PMC11239650
- DOI: 10.1038/s41597-024-03581-w
A near complete genome assembly of the East Friesian sheep genome
Erratum in
-
Author Correction: A near complete genome assembly of the East Friesian sheep genome.Sci Data. 2024 Aug 29;11(1):947. doi: 10.1038/s41597-024-03755-6. Sci Data. 2024. PMID: 39209856 Free PMC article. No abstract available.
Abstract
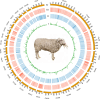
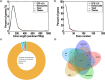
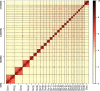
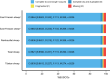
Advancements in sequencing have enabled the assembly of numerous sheep genomes, significantly advancing our understanding of the link between genetic variation and phenotypic traits. However, the genome of East Friesian sheep (Ostfriesisches Milchschaf), a key high-yield milk breed, remains to be fully assembled. Here, we constructed a near-complete and gap-free East Friesian genome assembly using PacBio HiFi, ultra-long ONT and Hi-C sequencing. The resulting genome assembly spans approximately 2.96 Gb, with a contig N50 length of 104.1 Mb and only 164 unplaced sequences. Remarkably, our assembly has captured 41 telomeres and 24 centromeres. The assembled sequence is of high quality on completeness (BUSCO score: 97.1%) and correctness (QV: 69.1). In addition, a total of 24,580 protein-coding genes were predicted, of which 97.2% (23,891) carried at least one conserved functional domain. Collectively, this assembly provides not only a near T2T gap-free genome, but also provides a valuable genetic resource for comparative genome studies of sheep and will serve as an important tool for the sheep research community.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Nguyen, Q. V. et al. Supplementing grazing dairy ewes with plant-derived oil and rumen-protected EPA+DHA pellets enhances health-beneficial n-3 long-chain polyunsaturated fatty acids in sheep milk. European Journal of Lipid Science and Technology120, 1700256 (2018). 10.1002/ejlt.201700256 - DOI
-
- Afolayan, R. A. et al. Genetic evaluation of crossbred lamb production. 3. Growth and carcass performance of second-cross lambs. Australian Journal of Agricultural Research58, 5 (2007). 10.1071/AR06310 - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

