Development and evaluation of a chronic kidney disease risk prediction model using random forest
- PMID: 38993480
- PMCID: PMC11236722
- DOI: 10.3389/fgene.2024.1409755
Development and evaluation of a chronic kidney disease risk prediction model using random forest
Abstract
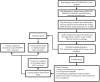
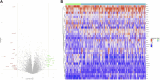
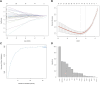
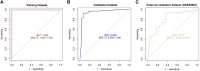
This research aims to advance the detection of Chronic Kidney Disease (CKD) through a novel gene-based predictive model, leveraging recent breakthroughs in gene sequencing. We sourced and merged gene expression profiles of CKD-affected renal tissues from the Gene Expression Omnibus (GEO) database, classifying them into two sets for training and validation in a 7:3 ratio. The training set included 141 CKD and 33 non-CKD specimens, while the validation set had 60 and 14, respectively. The disease risk prediction model was constructed using the training dataset, while the validation dataset confirmed the model's identification capabilities. The development of our predictive model began with evaluating differentially expressed genes (DEGs) between the two groups. We isolated six genes using Lasso and random forest (RF) methods-DUSP1, GADD45B, IFI44L, IFI30, ATF3, and LYZ-which are critical in differentiating CKD from non-CKD tissues. We refined our random forest (RF) model through 10-fold cross-validation, repeated five times, to optimize the mtry parameter. The performance of our model was robust, with an average AUC of 0.979 across the folds, translating to a 91.18% accuracy. Validation tests further confirmed its efficacy, with a 94.59% accuracy and an AUC of 0.990. External validation using dataset GSE180394 yielded an AUC of 0.913, 89.83% accuracy, and a sensitivity rate of 0.889, underscoring the model's reliability. In summary, the study identified critical genetic biomarkers and successfully developed a novel disease risk prediction model for CKD. This model can serve as a valuable tool for CKD disease risk assessment and contribute significantly to CKD identification.
Keywords: CKD; biomarkers; chronic kidney disease; computational genomics and proteomics; differentially expressed genes (DEGs); disease risk prediction algorithm; random forest..
Copyright © 2024 Mendapara.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Establishment and analysis of a disease risk prediction model for the systemic lupus erythematosus with random forest.Front Immunol. 2022 Nov 1;13:1025688. doi: 10.3389/fimmu.2022.1025688. eCollection 2022. Front Immunol. 2022. PMID: 36405750 Free PMC article.
-
Establishment and analysis of a novel diagnostic model for systemic juvenile idiopathic arthritis based on machine learning.Pediatr Rheumatol Online J. 2024 Jan 19;22(1):18. doi: 10.1186/s12969-023-00949-x. Pediatr Rheumatol Online J. 2024. PMID: 38243323 Free PMC article.
-
ESKD Risk Prediction Model in a Multicenter Chronic Kidney Disease Cohort in China: A Derivation, Validation, and Comparison Study.J Clin Med. 2023 Feb 14;12(4):1504. doi: 10.3390/jcm12041504. J Clin Med. 2023. PMID: 36836039 Free PMC article.
-
Development and validation of a machine learning-based predictive model for assessing the 90-day prognostic outcome of patients with spontaneous intracerebral hemorrhage.J Transl Med. 2024 Mar 4;22(1):236. doi: 10.1186/s12967-024-04896-3. J Transl Med. 2024. PMID: 38439097 Free PMC article.
-
Assessment of Renal Fibrosis in Patients With Chronic Kidney Disease Using Shear Wave Elastography and Clinical Features: A Random Forest Approach.Ultrasound Med Biol. 2023 Jul;49(7):1665-1671. doi: 10.1016/j.ultrasmedbio.2023.03.024. Epub 2023 Apr 25. Ultrasound Med Biol. 2023. PMID: 37105772
Cited by
-
The clinical prediction model to distinguish between colonization and infection by Klebsiella pneumoniae.Front Microbiol. 2025 Jan 23;15:1508030. doi: 10.3389/fmicb.2024.1508030. eCollection 2024. Front Microbiol. 2025. PMID: 39917270 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

