Comparative genome analyses of clinical and non-clinical Clostridioides difficile strains
- PMID: 38993487
- PMCID: PMC11238072
- DOI: 10.3389/fmicb.2024.1404491
Comparative genome analyses of clinical and non-clinical Clostridioides difficile strains
Abstract
The pathogenic bacterium Clostridioides difficile is a worldwide health burden with increasing morbidity, mortality and antibiotic resistances. Therefore, extensive research efforts are made to unravel its virulence and dissemination. One crucial aspect for C. difficile is its mobilome, which for instance allows the spread of antibiotic resistance genes (ARG) or influence strain virulence. As a nosocomial pathogen, the majority of strains analyzed originated from clinical environments and infected individuals. Nevertheless, C. difficile can also be present in human intestines without disease development or occur in diverse environmental habitats such as puddle water and soil, from which several strains could already be isolated. We therefore performed comprehensive genome comparisons of closely related clinical and non-clinical strains to identify the effects of the clinical background. Analyses included the prediction of virulence factors, ARGs, mobile genetic elements (MGEs), and detailed examinations of the pan genome. Clinical-related trends were thereby observed. While no significant differences were identified in fundamental C. difficile virulence factors, the clinical strains carried more ARGs and MGEs, and possessed a larger accessory genome. Detailed inspection of accessory genes revealed higher abundance of genes with unknown function, transcription-associated, or recombination-related activity. Accessory genes of these functions were already highlighted in other studies in association with higher strain virulence. This specific trend might allow the strains to react more efficiently on changing environmental conditions in the human host such as emerging stress factors, and potentially increase strain survival, colonization, and strain virulence. These findings indicated an adaptation of the strains to the clinical environment. Further, implementation of the analysis results in pairwise genome comparisons revealed that the majority of these accessory genes were encoded on predicted MGEs, shedding further light on the mobile genome of C. difficile. We therefore encourage the inclusion of non-clinical strains in comparative analyses.
Keywords: Clostridioide difficile; clinical; genome comparison; mobile genetic element; non-clinical; virulence.
Copyright © 2024 Schüler, Riedel, Overmann, Daniel and Poehlein.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
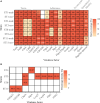

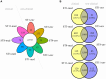





References
-
- Alcock B. P., Huynh W., Chalil R., Smith K. W., Raphenya A. R., Wlodarski M. A., et al. (2023). CARD 2023: expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res. 51, D690–D699. doi: 10.1093/nar/gkac920, PMID: - DOI - PMC - PubMed
-
- Alkudmani Z. S. B. (2018). The identification and characterization of novel haemolysin genes from Clostridium difficile: University College London.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

