Negative Regulation of CPSF6 Suppresses the Warburg Effect and Angiogenesis Leading to Tumor Progression Via c-Myc Signaling Network: Potential Therapeutic Target for Liver Cancer Therapy
- PMID: 38993554
- PMCID: PMC11234225
- DOI: 10.7150/ijbs.93462
Negative Regulation of CPSF6 Suppresses the Warburg Effect and Angiogenesis Leading to Tumor Progression Via c-Myc Signaling Network: Potential Therapeutic Target for Liver Cancer Therapy
Abstract
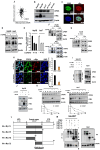
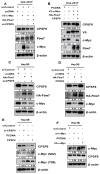
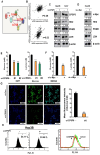
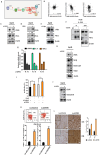
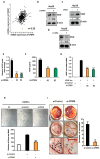
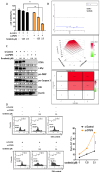
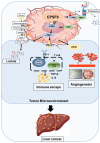
In this study, we explored the oncogenic mechanism of cleavage and polyadenylation-specific factor 6 (CPSF6) in hepatocellular carcinoma (HCC). CPSF6 was overexpressed in HCC tissues with poor survival rates compared to normal tissues. Hence, CPSF6 depletion suppressed cell viability and colony formation, induced apoptosis via PARP cleavage, and increased the sub-G1 population of Hep3B and Huh7 cells. In addition, CPSF6 enhanced the stability of c-Myc via their binding through nuclear co-localization by binding to c-Myc at the site of 258-360. Furthermore, c-Myc degradation by CPSF6 depletion was disturbed by FBW7 depletion or treatment with the proteasomal inhibitor MG132. Additionally, CPSF6 depletion suppressed the Warburg effect by inhibiting glucose, HK2, PKM2, LDH, and lactate; showed a synergistic effect with Sorafenib in Hep3B cells; and inhibited angiogenesis by tube formation and CAM assays, along with decreased expression and production of vascular endothelial growth factor (VEGF). Notably, CPSF6 depletion attenuated PD-L1 expression and increased Granzyme B levels, along with an increase in the percentage of CD4/CD8 cells in the splenocytes of BALB/c nude mice bearing Hep3B cells. Consistently, immunohistochemistry showed that CPSF6 depletion reduced the growth of Hep3B cells in BALB/c mice in orthotopic and xenograft tumor models by inhibiting tumor microenvironment-associated proteins. Overall, these findings suggest that CPSF6 enhances the Warburg effect for immune escape and angiogenesis, leading to cancer progression via c-Myc, mediated by the HK, PD-L1, and VEGF networks, with synergistic potential with sorafenib as a molecular target for liver cancer therapy.
Keywords: CPSF6; Warburg effect; angiogenesis; c-Myc; hepatocellular carcinoma.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures


References
-
- Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S. et al. Hepatocellular carcinoma. Nature reviews Disease primers. 2021;7:6. - PubMed
-
- Li H. Angiogenesis in the progression from liver fibrosis to cirrhosis and hepatocelluar carcinoma. Expert review of gastroenterology & hepatology. 2021;15:217–33. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

