TRIM50 inhibits glycolysis and the malignant progression of gastric cancer by ubiquitinating PGK1
- PMID: 38993561
- PMCID: PMC11234210
- DOI: 10.7150/ijbs.97091
TRIM50 inhibits glycolysis and the malignant progression of gastric cancer by ubiquitinating PGK1
Abstract
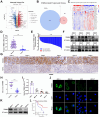
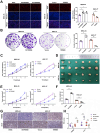
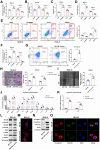
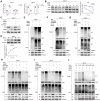
Ubiquitination plays a pivotal regulatory role in tumor progression. Among the components of the ubiquitin-proteasome system (UPS), ubiquitin-protein ligase E3 has emerged as a key molecule. Nevertheless, the biological functions of E3 ubiquitin ligases and their potential mechanisms orchestrating glycolysis in gastric cancer (GC) remain to be elucidated. In this study, we conducted a comprehensive transcriptomic analysis to identify the core E3 ubiquitin ligases in GC, followed by extensive validation of the expression patterns and clinical significance of Tripartite motif-containing 50 (TRIM50) both in vitro and in vivo. Remarkably, we found that TRIM50 was downregulated in GC tissues, associated with malignant progression and poor patient survival. Functionally, overexpression of TRIM50 suppressed GC cell proliferation and indirectly mitigated the invasion and migration of GC cells by inhibiting the M2 polarization of tumor-associated macrophages (TAMs). Mechanistically, TRIM50 inhibited the glycolytic pathway by ubiquitinating Phosphoglycerate Kinase 1 (PGK1), thereby directly suppressing GC cell proliferation. Simultaneously, the reduction in lactate led to diminished M2 polarization of TAMs, indirectly inhibiting the invasion and migration of GC cells. Notably, the downregulation of TRIM50 in GC was mediated by the METTL3/YTHDF2 axis in an m6A-dependent manner. In our study, we definitively identified TRIM50 as a tumor suppressor gene (TSG) that effectively inhibits glycolysis and the malignant progression of GC by ubiquitinating PGK1, thus offering novel insights and promising targets for the diagnosis and treatment of GC.
Keywords: Gastric cancer; glycolysis; m6A; tumor-associated macrophages; ubiquitination.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures





References
-
- Siegel RL, Miller KD, Wagle NS. et al. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17–48. - PubMed
-
- Ajani JA, D'Amico TA, Bentrem DJ. et al. Gastric Cancer, Version 2.2022, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2022;20(2):167–192. - PubMed
-
- Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat Med. 2014;20(11):1242–1253. - PubMed
-
- Zheng N, Shabek N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu Rev Biochem. 2017;86:129–157. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

