O-GlcNAcylated RALY Contributes to Hepatocellular Carcinoma Cells Proliferation by Regulating USP22 mRNA Nuclear Export
- PMID: 38993567
- PMCID: PMC11234212
- DOI: 10.7150/ijbs.97397
O-GlcNAcylated RALY Contributes to Hepatocellular Carcinoma Cells Proliferation by Regulating USP22 mRNA Nuclear Export
Abstract
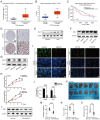
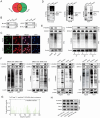
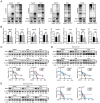
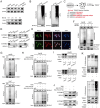
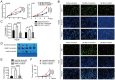
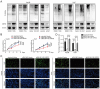
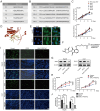
Hepatocellular carcinoma (HCC) is one of the most prevalent and deadly tumors; however, its pathogenic mechanism remains largely elusive. In-depth researches are needed to reveal the expression regulatory mechanisms and functions of the RNA-binding protein RALY in HCC. Here, we identify RALY as a highly expressed oncogenic factor that affects HCC cells proliferation both in vitro and in vivo. O-GlcNAcylation of RALY at Ser176 enhances its stability by protecting RALY from TRIM27-mediated ubiquitination, thus maintaining hyper-expression of the RALY protein. Mechanistically, RALY interacts with USP22 messenger RNA, as revealed by RNA immunoprecipitation, to increase their cytoplasmic localization and protein expression, thereby promoting the proliferation of HCC cells. Furthermore, we develop a novel RALY protein degrader based on peptide proteolysis-targeting chimeras, named RALY-PROTAC, which we chemically synthesize by linking a RALY-targeting peptide with the E3 ubiquitin ligase recruitment ligand pomalidomide. In conclusion, our findings demonstrate a novel mechanism by which O-GlcNAcylation/RALY/USP22 mRNA axis aggravates HCC cells proliferation. RALY-PROTACs as degraders of the RALY protein exhibit potential as therapeutic drugs for RALY-overexpressing HCC.
Keywords: O-GlcNAcylation; RALY; hepatocellular carcinoma; mRNA nuclear export; peptide proteolysis-targeting chimera.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures

References
-
- Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S. et al. Hepatocellular carcinoma. Nature Reviews Disease Primers. 2021;7:6. - PubMed
-
- Xu Y, Chou KC. Recent Progress in Predicting Posttranslational Modification Sites in Proteins. Current Topics in Medicinal Chemistry. 2016;16:591–603. - PubMed
-
- Shi J, Ruijtenbeek R, Pieters RJ. Demystifying O-GlcNAcylation: hints from peptide substrates. Glycobiology. 2018;28:814–24. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

