Fgf9 regulates bone marrow mesenchymal stem cell fate and bone-fat balance in osteoporosis by PI3K/AKT/Hippo and MEK/ERK signaling
- PMID: 38993574
- PMCID: PMC11234224
- DOI: 10.7150/ijbs.94863
Fgf9 regulates bone marrow mesenchymal stem cell fate and bone-fat balance in osteoporosis by PI3K/AKT/Hippo and MEK/ERK signaling
Abstract
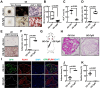
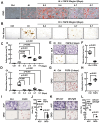
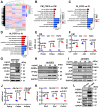
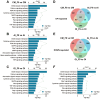
Bone-fat balance is crucial to maintain bone homeostasis. As common progenitor cells of osteoblasts and adipocytes, bone marrow mesenchymal stem cells (BMSCs) are delicately balanced for their differentiation commitment. However, the exact mechanisms governing BMSC cell fate are unclear. In this study, we discovered that fibroblast growth factor 9 (Fgf9), a cytokine expressed in the bone marrow niche, controlled bone-fat balance by influencing the cell fate of BMSCs. Histomorphology and cytodifferentiation analysis showed that Fgf9 loss-of-function mutation (S99N) notably inhibited bone marrow adipose tissue (BMAT) formation and alleviated ovariectomy-induced bone loss and BMAT accumulation in adult mice. Furthermore, in vitro and in vivo investigations demonstrated that Fgf9 altered the differentiation potential of BMSCs, shifting from osteogenesis to adipogenesis at the early stages of cell commitment. Transcriptomic and gene expression analyses demonstrated that FGF9 upregulated the expression of adipogenic genes while downregulating osteogenic gene expression at both mRNA and protein levels. Mechanistic studies revealed that FGF9, through FGFR1, promoted adipogenic gene expression via PI3K/AKT/Hippo pathways and inhibited osteogenic gene expression via MAPK/ERK pathway. This study underscores the crucial role of Fgf9 as a cytokine regulating the bone-fat balance in adult bone, suggesting that FGF9 is a potentially therapeutic target in the treatment of osteoporosis.
Keywords: Adipogenesis; Bone marrow adipose tissue; Bone-fate balance; Mesenchymal stem cells; Osteogenesis.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures




References
-
- Sims NA, Martin TJ. Chapter 4 - The osteoblast lineage: Its actions and communication mechanisms. In: Bilezikian JP, Martin TJ, Clemens TL, Rosen CJ, editors. Principles of Bone Biology (Fourth Edition): Academic Press. 2020. p. 89-110.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

