The CEBPB+ glioblastoma subcluster specifically drives the formation of M2 tumor-associated macrophages to promote malignancy growth
- PMID: 38994023
- PMCID: PMC11234274
- DOI: 10.7150/thno.93473
The CEBPB+ glioblastoma subcluster specifically drives the formation of M2 tumor-associated macrophages to promote malignancy growth
Abstract
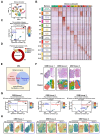
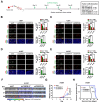
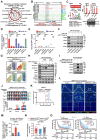
Rationale: The heterogeneity of tumor cells within the glioblastoma (GBM) microenvironment presents a complex challenge in curbing GBM progression. Understanding the specific mechanisms of interaction between different GBM cell subclusters and non-tumor cells is crucial. Methods: In this study, we utilized a comprehensive approach integrating glioma single-cell and spatial transcriptomics. This allowed us to examine the molecular interactions and spatial localization within GBM, focusing on a specific tumor cell subcluster, GBM subcluster 6, and M2-type tumor-associated macrophages (M2 TAMs). Results: Our analysis revealed a significant correlation between a specific tumor cell subcluster, GBM cluster 6, and M2-type TAMs. Further in vitro and in vivo experiments demonstrated the specific regulatory role of the CEBPB transcriptional network in GBM subcluster 6, which governs its tumorigenicity, recruitment of M2 TAMs, and polarization. This regulation involves molecules such as MCP1 for macrophage recruitment and the SPP1-Integrin αvβ1-Akt signaling pathway for M2 polarization. Conclusion: Our findings not only deepen our understanding of the formation of M2 TAMs, particularly highlighting the differential roles played by heterogeneous cells within GBM in this process, but also provided new insights for effectively controlling the malignant progression of GBM.
Keywords: CEBPB+ glioblastoma subcluster; Glioblastoma microenvironment; M2 Tumor-associated macrophages; SPP1-Integrin αvβ1-Akt axis; Single cell sequencing; Spatial transcriptome.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures





References
-
- Alexander BM, Cloughesy TF. Adult Glioblastoma. J Clin Oncol. 2017;35:2402–9. - PubMed
-
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJB, Janzer RC. et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–66. - PubMed
-
- Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

