Microglial circ-UBE2K exacerbates depression by regulating parental gene UBE2K via targeting HNRNPU
- PMID: 38994030
- PMCID: PMC11234284
- DOI: 10.7150/thno.96890
Microglial circ-UBE2K exacerbates depression by regulating parental gene UBE2K via targeting HNRNPU
Abstract
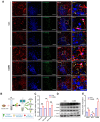
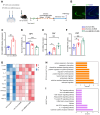
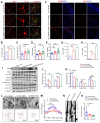
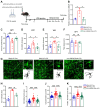
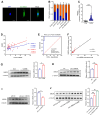
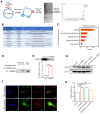
Background: Knowledge about the pathogenesis of depression and treatments for this disease are lacking. Epigenetics-related circRNAs are likely involved in the mechanism of depression and have great potential as treatment targets, but their mechanism of action is still unclear. Methods: Circular RNA UBE2K (circ-UBE2K) was screened from peripheral blood of patients with major depressive disorder (MDD) and brain of depression model mice through high-throughput sequencing. Microinjection of circ-UBE2K overexpression lentivirus and adeno-associated virus for interfering with microglial circ-UBE2K into the mouse hippocampus was used to observe the role of circ-UBE2K in MDD. Sucrose preference, forced swim, tail suspension and open filed tests were performed to evaluate the depressive-like behaviors of mice. Immunofluorescence and Western blotting analysis of the effects of circ-UBE2K on microglial activation and immune inflammation. Pull-down-mass spectrometry assay, RNA immunoprecipitation (RIP) test and fluorescence in situ hybridization (FISH) were used to identify downstream targets of circ-UBE2K/ HNRNPU (heterogeneous nuclear ribonucleoprotein U) axis. Results: In this study, through high-throughput sequencing and large-scale screening, we found that circ-UBE2K levels were significantly elevated both in the peripheral blood of patients with MDD and in the brains of depression model mice. Functionally, circ-UBE2K-overexpressing mice exhibited worsened depression-like symptoms, elevated brain inflammatory factor levels, and abnormal microglial activation. Knocking down circ-UBE2K mitigated these changes. Mechanistically, we found that circ-UBE2K binds to heterogeneous nuclear ribonucleoprotein U (HNRNPU) to form a complex that upregulates the expression of the parental gene ubiquitin conjugating enzyme E2 K (UBE2K), leading to abnormal microglial activation and neuroinflammation and promoting the occurrence and development of depression. Conclusions: The findings of the present study revealed that the expression of circUBE2K, which combines with HNRNPU to form the circUBE2K/HNRNPU complex, is increased in microglia after external stress, thus regulating the expression of the parental gene UBE2K and mediating the abnormal activation of microglia to induce neuroinflammation, promoting the development of MDD. These results indicate that circ-UBE2K plays a newly discovered role in the pathogenesis of depression.
Keywords: circular RNA UBE2K (circ-UBE2K); heterogeneous nuclear ribonucleoprotein U (HNRNPU); major depressive disorder (MDD); microglial activation; neuroinflammation.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

