Action of circulating and infiltrating B cells in the immune microenvironment of colorectal cancer by single-cell sequencing analysis
- PMID: 38994150
- PMCID: PMC11236258
- DOI: 10.4251/wjgo.v16.i6.2683
Action of circulating and infiltrating B cells in the immune microenvironment of colorectal cancer by single-cell sequencing analysis
Abstract
Background: The complexity of the immune microenvironment has an impact on the treatment of colorectal cancer (CRC), one of the most prevalent malignancies worldwide. In this study, multi-omics and single-cell sequencing techniques were used to investigate the mechanism of action of circulating and infiltrating B cells in CRC. By revealing the heterogeneity and functional differences of B cells in cancer immunity, we aim to deepen our understanding of immune regulation and provide a scientific basis for the development of more effective cancer treatment strategies.
Aim: To explore the role of circulating and infiltrating B cell subsets in the immune microenvironment of CRC, explore the potential driving mechanism of B cell development, analyze the interaction between B cells and other immune cells in the immune microenvironment and the functions of communication molecules, and search for possible regulatory pathways to promote the anti-tumor effects of B cells.
Methods: A total of 69 paracancer (normal), tumor and peripheral blood samples were collected from 23 patients with CRC from The Cancer Genome Atlas database (https://portal.gdc.cancer.gov/). After the immune cells were sorted by multicolor flow cytometry, the single cell transcriptome and B cell receptor group library were sequenced using the 10X Genomics platform, and the data were analyzed using bioinformatics tools such as Seurat. The differences in the number and function of B cell infiltration between tumor and normal tissue, the interaction between B cell subsets and T cells and myeloid cell subsets, and the transcription factor regulatory network of B cell subsets were explored and analyzed.
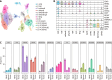
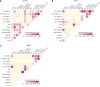
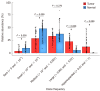
Results: Compared with normal tissue, the infiltrating number of CD20+B cell subsets in tumor tissue increased significantly. Among them, germinal center B cells (GCB) played the most prominent role, with positive clone expansion and heavy chain mutation level increasing, and the trend of differentiation into memory B cells increased. However, the number of plasma cells in the tumor microenvironment decreased significantly, and the plasma cells secreting IgA antibodies decreased most obviously. In addition, compared with the immune microenvironment of normal tissues, GCB cells in tumor tissues became more closely connected with other immune cells such as T cells, and communication molecules that positively regulate immune function were significantly enriched.
Conclusion: The role of GCB in CRC tumor microenvironment is greatly enhanced, and its affinity to tumor antigen is enhanced by its significantly increased heavy chain mutation level. Meanwhile, GCB has enhanced its association with immune cells in the microenvironment, which plays a positive anti-tumor effect.
Keywords: Colorectal cancer; Immune microenvironment; Infiltrating B cells; Multi-omics analysis; Single-cell sequencing analysis.
©The Author(s) 2024. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: The authors declare no conflicts of interest.
Figures






Similar articles
-
Integration of single-cell RNA sequencing and bulk RNA transcriptome sequencing reveals a heterogeneous immune landscape and pivotal cell subpopulations associated with colorectal cancer prognosis.Front Immunol. 2023 Aug 22;14:1184167. doi: 10.3389/fimmu.2023.1184167. eCollection 2023. Front Immunol. 2023. PMID: 37675100 Free PMC article.
-
Mapk14 is a Prognostic Biomarker and Correlates with the Clinicopathological Features and Immune Infiltration of Colorectal Cancer.Front Cell Dev Biol. 2022 Jan 24;10:817800. doi: 10.3389/fcell.2022.817800. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35141222 Free PMC article.
-
Research on the Regulatory Mechanism of Ginseng on the Tumor Microenvironment of Colorectal Cancer based on Network Pharmacology and Bioinformatics Validation.Curr Comput Aided Drug Des. 2024;20(5):486-500. doi: 10.2174/1573409919666230607103721. Curr Comput Aided Drug Des. 2024. PMID: 37287284
-
Colorectal Cancer-Infiltrating Regulatory T Cells: Functional Heterogeneity, Metabolic Adaptation, and Therapeutic Targeting.Front Immunol. 2022 Jul 8;13:903564. doi: 10.3389/fimmu.2022.903564. eCollection 2022. Front Immunol. 2022. PMID: 35874729 Free PMC article. Review.
-
Unravelling the heterogeneity and dynamic relationships of tumor-infiltrating T cells by single-cell RNA sequencing analysis.J Leukoc Biol. 2020 Jun;107(6):917-932. doi: 10.1002/JLB.6MR0320-234R. Epub 2020 Apr 9. J Leukoc Biol. 2020. PMID: 32272497 Free PMC article. Review.
Cited by
-
Causal relationship between immune cell signatures and colorectal cancer: a bi-directional, two-sample mendelian randomization study.BMC Cancer. 2025 Mar 3;25(1):387. doi: 10.1186/s12885-025-13576-4. BMC Cancer. 2025. PMID: 40033246 Free PMC article.
-
Exploring the role of neutrophil extracellular traps in colorectal cancer: Insights from single-cell sequencing.World J Gastrointest Oncol. 2025 Jul 15;17(7):107589. doi: 10.4251/wjgo.v17.i7.107589. World J Gastrointest Oncol. 2025. PMID: 40697221 Free PMC article. Review.
References
-
- Zhang L, Li Z, Skrzypczynska KM, Fang Q, Zhang W, O'Brien SA, He Y, Wang L, Zhang Q, Kim A, Gao R, Orf J, Wang T, Sawant D, Kang J, Bhatt D, Lu D, Li CM, Rapaport AS, Perez K, Ye Y, Wang S, Hu X, Ren X, Ouyang W, Shen Z, Egen JG, Zhang Z, Yu X. Single-Cell Analyses Inform Mechanisms of Myeloid-Targeted Therapies in Colon Cancer. Cell. 2020;181:442–459.e29. - PubMed
-
- Li J, Wu C, Hu H, Qin G, Wu X, Bai F, Zhang J, Cai Y, Huang Y, Wang C, Yang J, Luan Y, Jiang Z, Ling J, Wu Z, Chen Y, Xie Z, Deng Y. Remodeling of the immune and stromal cell compartment by PD-1 blockade in mismatch repair-deficient colorectal cancer. Cancer Cell. 2023;41:1152–1169.e7. - PubMed
-
- Joanito I, Wirapati P, Zhao N, Nawaz Z, Yeo G, Lee F, Eng CLP, Macalinao DC, Kahraman M, Srinivasan H, Lakshmanan V, Verbandt S, Tsantoulis P, Gunn N, Venkatesh PN, Poh ZW, Nahar R, Oh HLJ, Loo JM, Chia S, Cheow LF, Cheruba E, Wong MT, Kua L, Chua C, Nguyen A, Golovan J, Gan A, Lim WJ, Guo YA, Yap CK, Tay B, Hong Y, Chong DQ, Chok AY, Park WY, Han S, Chang MH, Seow-En I, Fu C, Mathew R, Toh EL, Hong LZ, Skanderup AJ, DasGupta R, Ong CJ, Lim KH, Tan EKW, Koo SL, Leow WQ, Tejpar S, Prabhakar S, Tan IB. Single-cell and bulk transcriptome sequencing identifies two epithelial tumor cell states and refines the consensus molecular classification of colorectal cancer. Nat Genet. 2022;54:963–975. - PMC - PubMed
-
- Wu Y, Yang S, Ma J, Chen Z, Song G, Rao D, Cheng Y, Huang S, Liu Y, Jiang S, Liu J, Huang X, Wang X, Qiu S, Xu J, Xi R, Bai F, Zhou J, Fan J, Zhang X, Gao Q. Spatiotemporal Immune Landscape of Colorectal Cancer Liver Metastasis at Single-Cell Level. Cancer Discov. 2022;12:134–153. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

