Gravidity influences distinct transcriptional profiles of maternal and fetal placental macrophages at term
- PMID: 38994356
- PMCID: PMC11237841
- DOI: 10.3389/fimmu.2024.1384361
Gravidity influences distinct transcriptional profiles of maternal and fetal placental macrophages at term
Abstract
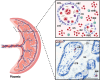
Introduction: Maternal intervillous monocytes (MIMs) and fetal Hofbauer cells (HBCs) are myeloid-derived immune cells at the maternal-fetal interface. Maternal reproductive history is associated with differential risk of pregnancy complications. The molecular phenotypes and roles of these distinct monocyte/macrophage populations and the influence of gravidity on these phenotypes has not been systematically investigated.
Methods: Here, we used RNA sequencing to study the transcriptional profiles of MIMs and HBCs in normal term pregnancies.
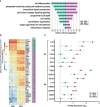
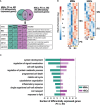
Results: Our analyses revealed distinct transcriptomes of MIMs and HBCs. Genes involved in differentiation and cell organization pathways were more highly expressed in MIMs vs. HBCs. In contrast, HBCs had higher expression of genes involved in inflammatory responses and cell surface receptor signaling. Maternal gravidity influenced monocyte programming, as expression of pro-inflammatory molecules was significantly higher in MIMs from multigravidae compared to primigravidae. In HBCs, multigravidae displayed enrichment of gene pathways involved in cell-cell signaling and differentiation.
Discussion: Our results demonstrated that MIMs and HBCs have highly divergent transcriptional signatures, reflecting their distinct origins, locations, functions, and roles in inflammatory responses. Furthermore, maternal gravidity influences the gene signatures of MIMs and HBCs, potentially modulating the interplay between tolerance and trained immunity. The phenomenon of reproductive immune memory may play a novel role in the differential susceptibility of primigravidae to pregnancy complications.
Keywords: Hofbauer cell; RNA-Seq; gravidity; monocyte; myeloid cells; placenta; pregnancy; transcriptomics.
Copyright © 2024 Ozarslan, Robinson, Buarpung, Kim, Ansbro, Akram, Montoya, Kamya, Kakuru, Dorsey, Rosenthal, Cheng, Feeney, Fisher and Gaw.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Update of
-
Distinct transcriptional profiles of maternal and fetal placental macrophages at term are associated with gravidity.bioRxiv [Preprint]. 2023 Sep 28:2023.09.25.559419. doi: 10.1101/2023.09.25.559419. bioRxiv. 2023. Update in: Front Immunol. 2024 Jun 26;15:1384361. doi: 10.3389/fimmu.2024.1384361. PMID: 37808856 Free PMC article. Updated. Preprint.
References
-
- Labarrere CA, DiCarlo HL, Bammerlin E, Hardin JW, Kim YM, Chaemsaithong P, et al. . Failure of physiologic transformation of spiral arteries, endothelial and trophoblast cell activation, and acute atherosis in the basal plate of the placenta. Am J Obstet Gynecol. (2017) 216:287.e1–287.e16. doi: 10.1016/j.ajog.2016.12.029 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

