Single-cell mapping of leukocyte immunoglobulin-like receptors in kidney transplant rejection
- PMID: 38994376
- PMCID: PMC11235271
- DOI: 10.3389/frtra.2022.952785
Single-cell mapping of leukocyte immunoglobulin-like receptors in kidney transplant rejection
Abstract
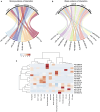
Leukocyte immunoglobulin-like receptors (LILRs) are a family of inhibitory or stimulatory receptors expressed by immune cell types belonging to both myeloid and lymphoid lineage. Several members of the LILR family recognize major histocompatibility complex class I and thus play important roles in a range of clinical situations including pregnancy. Moreover, paired immunoglobulin-like receptors (PIRs), the murine orthologs of LILRs, are implicated in experimental transplant allorecognition by monocytes and contribute to the induction of donor-specific monocyte-memory. After non-self recognition, activating PIRs are transiently overexpressed at the surface of monocytes and participate in donor-specific monocyte recruitment, leading to graft rejection in vivo. In the present study, we mapped LILR expression and also their respective reported ligands at single cell level in the renal allograft and circulating cells in the context of kidney transplant rejection. Recipient-derived monocytes were shown to infiltrate the donor tissue and to differentiate into macrophages. We thus also investigate LILR expression during in vitro monocyte-to-macrophage differentiation in order to characterize the myeloid population that directly contribute to allorecognition. Altogether our results emphasize non-classical monocytes and CD68+ M1 macrophages as key players in LILRs-ligand interaction in kidney transplantation.
Keywords: LILR; allorecognition; kidney transplantation; monocytes; single-cell RNA sequencing.
Copyright © 2022 Lamarthée, Genet, Cattin, Danger, Giral, Brouard, Van Loon, Callemeyn, Naesens, Anglicheau, Bonnotte, Legendre, Rebibou and Tinel.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Haas M, Loupy A, Lefaucheur C, Roufosse C, Glotz D, Seron D, et al. . The Banff 2017 kidney meeting report : revised diagnostic criteria for chronic active T cell – mediated rejection, antibody- mediated rejection, and prospects for integrative endpoints for next- generation clinical trials. Am J Transplant. (2018) 18:293–307. 10.1111/ajt.14625 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

