Distinctive serotypes of SARS-related coronaviruses defined by convalescent sera from unvaccinated individuals
- PMID: 38994526
- PMCID: PMC11238253
- DOI: 10.1016/j.hlife.2023.07.002
Distinctive serotypes of SARS-related coronaviruses defined by convalescent sera from unvaccinated individuals
Abstract
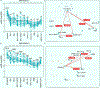
Multiple Omicron sub-lineages have emerged, with Omicron XBB and XBB.1.5 subvariants becoming the dominant variants globally at the time of this study. The key feature of new variants is their ability to escape humoral immunity despite the fact that there are limited genetic changes from their preceding variants. This raises the question of whether Omicron should be regarded as a separate serotype from viruses serologically clustered with the ancestral severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus. Here, we present cross-neutralization data based on a pseudovirus neutralization test using convalescent sera from naïve individuals who had recovered from primary infection by SARS-CoV-1 and SARS-CoV-2 strains/variants including the ancestral virus and variants Beta, Delta, Omicron BA.1, Omicron BA.2 and Omicron BA.5. The results revealed no significant cross-neutralization in any of the three-way testing for SARS-CoV-1, ancestral SARS-CoV-2 and SARS-CoV-2 Omicron subvariants. The data argue for the assignment of three distinct serotypes for the currently known human-infecting SARS-related coronaviruses.
Keywords: Omicron; SARS-CoV-1; SARS-CoV-2; convalescent sera; primary infection; serotype.
Conflict of interest statement
DECLARATION OF COMPETING INTERESTS The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Tan CW, Chia WN, Zhu F, Young BE, Chantasrisawad N, Hwa SH, et al. SARS-CoV-2 Omicron variant emerged under immune selection. Nat Microbiol 2022;7:1756–61. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
