Data Independent Acquisition Mass Spectrometry Enhanced Personalized Glycosylation Profiling of Haptoglobin in Hepatocellular Carcinoma
- PMID: 38994555
- PMCID: PMC11301664
- DOI: 10.1021/acs.jproteome.4c00227
Data Independent Acquisition Mass Spectrometry Enhanced Personalized Glycosylation Profiling of Haptoglobin in Hepatocellular Carcinoma
Abstract
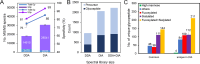
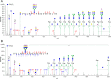
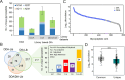
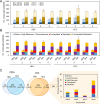
Aberrant glycosylation has gained significant interest for biomarker discovery. However, low detectability, complex glycan structures, and heterogeneity present challenges in glycoprotein assay development. Using haptoglobin (Hp) as a model, we developed an integrated platform combining functionalized magnetic nanoparticles and zwitterionic hydrophilic interaction liquid chromatography (ZIC-HILIC) for highly specific glycopeptide enrichment, followed by a data-independent acquisition (DIA) strategy to establish a deep cancer-specific Hp-glycosylation profile in hepatitis B virus (HBV, n = 5) and hepatocellular carcinoma (HCC, n = 5) patients. The DIA strategy established one of the deepest Hp-glycosylation landscapes (1029 glycopeptides, 130 glycans) across serum samples, including 54 glycopeptides exclusively detected in HCC patients. Additionally, single-shot DIA searches against a DIA-based spectral library outperformed the DDA approach by 2-3-fold glycopeptide coverage across patients. Among the four N-glycan sites on Hp (N-184, N-207, N-211, N-241), the total glycan type distribution revealed significantly enhanced detection of combined fucosylated-sialylated glycans, which were the most dominant glycoforms identified in HCC patients. Quantitation analysis revealed 48 glycopeptides significantly enriched in HCC (p < 0.05), including a hybrid monosialylated triantennary glycopeptide on the N-184 site with nearly none-to-all elevation to differentiate HCC from the HBV group (HCC/HBV ratio: 2462 ± 766, p < 0.05). In summary, DIA-MS presents an unbiased and comprehensive alternative for targeted glycoproteomics to guide discovery and validation of glyco-biomarkers.
Keywords: data-independent acquisition (DIA); glycoprotein; haptoglobin; hepatocellular carcinoma (HCC).
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Quantitative analysis of site-specific N-glycans on sera haptoglobin β chain in liver diseases.Acta Biochim Biophys Sin (Shanghai). 2013 Dec;45(12):1021-9. doi: 10.1093/abbs/gmt110. Epub 2013 Oct 8. Acta Biochim Biophys Sin (Shanghai). 2013. PMID: 24103369
-
Clinical glycoprotein mass spectrometry: The future of disease detection and monitoring.J Mass Spectrom. 2024 Sep;59(9):e5083. doi: 10.1002/jms.5083. J Mass Spectrom. 2024. PMID: 39162140 Review.
-
LC-MS/MS isomeric profiling of permethylated N-glycans derived from serum haptoglobin of hepatocellular carcinoma (HCC) and cirrhotic patients.Electrophoresis. 2017 Sep;38(17):2160-2167. doi: 10.1002/elps.201700025. Epub 2017 Jul 14. Electrophoresis. 2017. PMID: 28543513 Free PMC article.
-
An Integrated Strategy Using Predicted Spectral Library and Tandem Enrichment for Large-Scale Identification of Low-Abundance Proteins and Intact N-Glycopeptides in Human Plasma.Anal Chem. 2025 Jul 29;97(29):15855-15863. doi: 10.1021/acs.analchem.5c02002. Epub 2025 Jul 17. Anal Chem. 2025. PMID: 40674567
-
Current Advancements in Serum Protein Biomarkers for Hepatitis B Virus-Associated Hepatocyte Remodeling and Hepatocellular Carcinoma.Immun Inflamm Dis. 2025 Apr;13(4):e70171. doi: 10.1002/iid3.70171. Immun Inflamm Dis. 2025. PMID: 40192058 Free PMC article. Review.
Cited by
-
In-depth plasma N-glycoproteome profiling using narrow-window data-independent acquisition on the Orbitrap Astral mass spectrometer.Nat Commun. 2025 Mar 13;16(1):2497. doi: 10.1038/s41467-025-57916-1. Nat Commun. 2025. PMID: 40082474 Free PMC article.
References
-
- Liu Y. C.; Yen H. Y.; Chen C. Y.; Chen C. H.; Cheng P. F.; Juan Y. H.; Chen C. H.; Khoo K. H.; Yu C. J.; Yang P. C.; Hsu T. L.; Wong C. H. Sialylation and fucosylation of epidermal growth factor receptor suppress its dimerization and activation in lung cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2011, 108 (28), 11332–11337. 10.1073/pnas.1107385108. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

