p53R172H and p53R245W Hotspot Mutations Drive Distinct Transcriptomes in Mouse Mammary Tumors Through a Convergent Transcriptional Mediator
- PMID: 38994678
- PMCID: PMC11310746
- DOI: 10.1158/2767-9764.CRC-24-0128
p53R172H and p53R245W Hotspot Mutations Drive Distinct Transcriptomes in Mouse Mammary Tumors Through a Convergent Transcriptional Mediator
Abstract
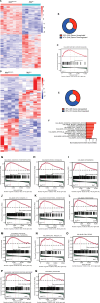
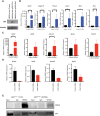
Aggressive breast cancers harbor TP53 missense mutations. Tumor cells with TP53 missense mutations exhibit enhanced growth and survival through transcriptional rewiring. To delineate how TP53 mutations in breast cancer contribute to tumorigenesis and progression in vivo, we created a somatic mouse model driven by mammary epithelial cell-specific expression of Trp53 mutations. Mice developed primary mammary tumors reflecting the human molecular subtypes of luminal A, luminal B, HER2-enriched, and triple-negative breast cancer with metastases. Transcriptomic analyses comparing MaPR172H/- or MaPR245W/- mammary tumors to MaP-/- tumors revealed (1) differences in cancer-associated pathways activated in both p53 mutants and (2) Nr5a2 as a novel transcriptional mediator of distinct pathways in p53 mutants. Meta-analyses of human breast tumors corroborated these results. In vitro assays demonstrate mutant p53 upregulates specific target genes that are enriched for Nr5a2 response elements in their promoters. Co-immunoprecipitation studies revealed p53R172H and p53R245W interact with Nr5a2. These findings implicate NR5A2 as a novel mediator of mutant p53 transcriptional activity in breast cancer.
Significance: Our findings implicate NR5A2 as a novel mediator of mutant p53 transcriptional activity in breast cancer. NR5A2 may be an important therapeutic target in hard-to-treat breast cancers such as endocrine-resistant tumors and metastatic triple-negative breast cancers harboring TP53 missense mutations.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
G. Lozano reports grants from NCI and CPRIT during the conduct of the study. No disclosures were reported by the other authors.
Figures




Similar articles
-
Somatic Trp53 mutations differentially drive breast cancer and evolution of metastases.Nat Commun. 2018 Sep 27;9(1):3953. doi: 10.1038/s41467-018-06146-9. Nat Commun. 2018. PMID: 30262850 Free PMC article.
-
Targeted Pten deletion plus p53-R270H mutation in mouse mammary epithelium induces aggressive claudin-low and basal-like breast cancer.Breast Cancer Res. 2016 Jan 19;18(1):9. doi: 10.1186/s13058-015-0668-y. Breast Cancer Res. 2016. PMID: 26781438 Free PMC article.
-
Triple-negative breast tumors are dependent on mutant p53 for growth and survival.Proc Natl Acad Sci U S A. 2023 Aug 22;120(34):e2308807120. doi: 10.1073/pnas.2308807120. Epub 2023 Aug 14. Proc Natl Acad Sci U S A. 2023. PMID: 37579145 Free PMC article.
-
p53 in breast cancer subtypes and new insights into response to chemotherapy.Breast. 2013 Aug;22 Suppl 2:S27-9. doi: 10.1016/j.breast.2013.07.005. Breast. 2013. PMID: 24074787 Review.
-
Mouse models of PIK3CA mutations: one mutation initiates heterogeneous mammary tumors.FEBS J. 2013 Jun;280(12):2758-65. doi: 10.1111/febs.12175. Epub 2013 Mar 1. FEBS J. 2013. PMID: 23384338 Review.
Cited by
-
Genomic Differences and Distinct TP53 Mutation Site-Linked Chemosensitivity in Early- and Late-Onset Gastric Cancer.Cancer Med. 2025 Apr;14(8):e70793. doi: 10.1002/cam4.70793. Cancer Med. 2025. PMID: 40249206 Free PMC article.
References
-
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. . Molecular portraits of human breast tumours. Nature 2000;406:747–52. - PubMed
-
- Hoch RV, Thompson DA, Baker RJ, Weigel RJ. GATA-3 is expressed in association with estrogen receptor in breast cancer. Int J Cancer 1999;84:122–8. - PubMed
-
- Rastelli F, Crispino S. Factors predictive of response to hormone therapy in breast cancer. Tumori 2008;94:370–83. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

