Role of LMO7 in cancer (Review)
- PMID: 38994754
- PMCID: PMC11267500
- DOI: 10.3892/or.2024.8776
Role of LMO7 in cancer (Review)
Abstract
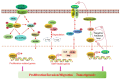
Cancer constitutes a multifaceted ailment characterized by the dysregulation of numerous genes and pathways. Among these, LIM domain only 7 (LMO7) has emerged as a significant player in various cancer types, garnering substantial attention for its involvement in tumorigenesis and cancer progression. This review endeavors to furnish a comprehensive discourse on the functional intricacies and mechanisms of LMO7 in cancer, with a particular emphasis on its potential as both a therapeutic target and prognostic indicator. It delves into the molecular attributes of LMO7, its implications in cancer etiology and the underlying mechanisms propelling its oncogenic properties. Furthermore, it underscores the extant challenges and forthcoming prospects in targeting LMO7 for combating cancer.
Keywords: LIM domain only 7; cancer; tumorigenesis.
Conflict of interest statement
The authors declare that they have no competing interests.
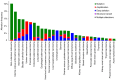
Figures
References
-
- Wang X, Chen G, Zhang Y, Ghareeb WM, Yu Q, Zhu H, Lu X, Huang Y, Huang S, Hou D, Chi P. The impact of circumferential tumour location on the clinical outcome of rectal cancer patients managed with neoadjuvant chemoradiotherapy followed by total mesorectal excision. Eur J Surg Oncol. 2020;46:1118–1123. doi: 10.1016/j.ejso.2020.07.024. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

