Global Transcriptomic and Characteristics Comparisons between Mouse Fetal Liver and Bone Marrow Definitive Erythropoiesis
- PMID: 38995000
- PMCID: PMC11240549
- DOI: 10.3390/cells13131149
Global Transcriptomic and Characteristics Comparisons between Mouse Fetal Liver and Bone Marrow Definitive Erythropoiesis
Abstract
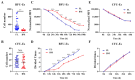
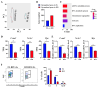
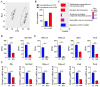
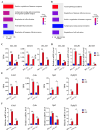
Erythropoiesis occurs first in the yolk sac as a transit "primitive" form, then is gradually replaced by the "definitive" form in the fetal liver (FL) during fetal development and in the bone marrow (BM) postnatally. While it is well known that differences exist between primitive and definitive erythropoiesis, the similarities and differences between FL and BM definitive erythropoiesis have not been studied. Here we performed comprehensive comparisons of erythroid progenitors and precursors at all maturational stages sorted from E16.5 FL and adult BM. We found that FL cells at all maturational stages were larger than their BM counterparts. We further found that FL BFU-E cells divided at a faster rate and underwent more cell divisions than BM BFU-E. Transcriptome comparison revealed that genes with increased expression in FL BFU-Es were enriched in cell division. Interestingly, the expression levels of glucocorticoid receptor Nr3c1, Myc and Myc downstream target Ccna2 were significantly higher in FL BFU-Es, indicating the role of the Nr3c1-Myc-Ccna2 axis in the enhanced proliferation/cell division of FL BFU-E cells. At the CFU-E stage, the expression of genes associated with hemoglobin biosynthesis were much higher in FL CFU-Es, indicating more hemoglobin production. During terminal erythropoiesis, overall temporal patterns in gene expression were conserved between the FL and BM. While biological processes related to translation, the tricarboxylic acid cycle and hypoxia response were upregulated in FL erythroblasts, those related to antiviral signal pathway were upregulated in BM erythroblasts. Our findings uncovered previously unrecognized differences between FL and BM definitive erythropoiesis and provide novel insights into erythropoiesis.
Keywords: bone marrow; cell cycle; definitive erythropoiesis; fetal liver; transcriptome analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

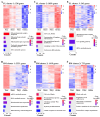

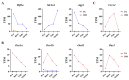
Similar articles
-
Functional Analysis of Erythroid Progenitors by Colony-Forming Assays.Methods Mol Biol. 2018;1698:117-132. doi: 10.1007/978-1-4939-7428-3_7. Methods Mol Biol. 2018. PMID: 29076087
-
Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse.Development. 1999 Nov;126(22):5073-84. doi: 10.1242/dev.126.22.5073. Development. 1999. PMID: 10529424
-
Comprehensive Characterization and Global Transcriptome Analysis of Human Fetal Liver Terminal Erythropoiesis.Genomics Proteomics Bioinformatics. 2023 Dec;21(6):1117-1132. doi: 10.1016/j.gpb.2023.07.001. Epub 2023 Aug 30. Genomics Proteomics Bioinformatics. 2023. PMID: 37657739 Free PMC article.
-
Erythropoiesis in the mammalian embryo.Exp Hematol. 2024 Aug;136:104283. doi: 10.1016/j.exphem.2024.104283. Epub 2024 Jul 22. Exp Hematol. 2024. PMID: 39048071 Review.
-
Ontogeny of erythropoiesis in the mammalian embryo.Curr Top Dev Biol. 2008;82:1-22. doi: 10.1016/S0070-2153(07)00001-4. Curr Top Dev Biol. 2008. PMID: 18282515 Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

