Mitonuclear compatibility is maintained despite relaxed selection on male mitochondrial DNA in bivalves with doubly uniparental inheritance
- PMID: 38995057
- PMCID: PMC11519007
- DOI: 10.1093/evolut/qpae108
Mitonuclear compatibility is maintained despite relaxed selection on male mitochondrial DNA in bivalves with doubly uniparental inheritance
Abstract
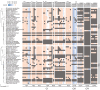
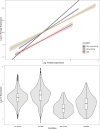
Mitonuclear coevolution is common in eukaryotes, but bivalve lineages that have doubly uniparental inheritance (DUI) of mitochondria may be an interesting example. In this system, females transmit mtDNA (F mtDNA) to all offspring, while males transmit a different mtDNA (M mtDNA) solely to their sons. Molecular evolution and functional data suggest oxidative phosphorylation (OXPHOS) genes encoded in M mtDNA evolve under relaxed selection due to their function being limited to sperm only (vs. all other tissues for F mtDNA). This has led to the hypothesis that mitonuclear coevolution is less important for M mtDNA. Here, we use comparative phylogenetics, transcriptomics, and proteomics to understand mitonuclear interactions in DUI bivalves. We found nuclear OXPHOS proteins coevolve and maintain compatibility similarly with both F and M mtDNA OXPHOS proteins. Mitochondrial recombination did not influence mitonuclear compatibility and nuclear-encoded OXPHOS genes were not upregulated in tissues with M mtDNA to offset dysfunction. Our results support that selection maintains mitonuclear compatibility with F and M mtDNA despite relaxed selection on M mtDNA. Strict sperm transmission, lower effective population size, and higher mutation rates may explain the evolution of M mtDNA. Our study highlights that mitonuclear coevolution and compatibility may be broad features of eukaryotes.
Keywords: bivalvia; evolutionary rate covariation; molecular evolution; phylogenetics; proteomics; transcriptomics.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Society for the Study of Evolution (SSE). All rights reserved. For commercial re-use, please contact reprints@oup.com for reprints and translation rights for reprints. All other permissions can be obtained through our RightsLink service via the Permissions link on the article page on our site—for further information please contact journals.permissions@oup.com.
Conflict of interest statement
The authors do not declare any conflicts of interest.
Figures


Similar articles
-
Mitonuclear Coevolution in Bumblebees (Bombus): Genomic Signatures and Its Role in Climatic Niche Adaptation.Genome Biol Evol. 2025 Jul 3;17(7):evaf123. doi: 10.1093/gbe/evaf123. Genome Biol Evol. 2025. PMID: 40509913 Free PMC article.
-
Relaxed selection on male mitochondrial genes in DUI bivalves eases the need for mitonuclear coevolution.J Evol Biol. 2021 Nov;34(11):1722-1736. doi: 10.1111/jeb.13931. Epub 2021 Sep 29. J Evol Biol. 2021. PMID: 34533872 Free PMC article.
-
A tale of two paths: The evolution of mitochondrial recombination in bivalves with doubly uniparental inheritance.J Hered. 2023 May 25;114(3):199-206. doi: 10.1093/jhered/esad004. J Hered. 2023. PMID: 36897956 Free PMC article.
-
Gender differences in the context of interventions for improving health literacy in migrants: a qualitative evidence synthesis.Cochrane Database Syst Rev. 2024 Dec 12;12(12):CD013302. doi: 10.1002/14651858.CD013302.pub2. Cochrane Database Syst Rev. 2024. PMID: 39665382
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
Cited by
-
ERCnet: Phylogenomic Prediction of Interaction Networks in the Presence of Gene Duplication.Mol Biol Evol. 2025 Apr 30;42(5):msaf089. doi: 10.1093/molbev/msaf089. Mol Biol Evol. 2025. PMID: 40247660 Free PMC article.
References
-
- Amil-Ruiz, F., Herruzo-Ruiz, A. M., Fuentes-Almagro, C., Baena-Angulo, C., Jiménez-Pastor, J. M., Blasco, J., Michán, C., & Michán, C. (2021). Constructing a de novo transcriptome and a reference proteome for the bivalve Scrobicularia plana: Comparative analysis of different assembly strategies and proteomic analysis. Genomics, 113(3), 1543–1553. 10.1016/j.ygeno.2021.03.025 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
