Robust reprogramming of glia into neurons by inhibition of Notch signaling and nuclear factor I (NFI) factors in adult mammalian retina
- PMID: 38996013
- PMCID: PMC11244444
- DOI: 10.1126/sciadv.adn2091
Robust reprogramming of glia into neurons by inhibition of Notch signaling and nuclear factor I (NFI) factors in adult mammalian retina
Abstract
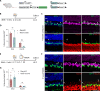
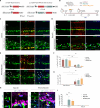
Generation of neurons through direct reprogramming has emerged as a promising therapeutic approach for treating neurodegenerative diseases. In this study, we present an efficient method for reprogramming retinal glial cells into neurons. By suppressing Notch signaling by disrupting either Rbpj or Notch1/2, we induced mature Müller glial cells to reprogram into bipolar- and amacrine-like neurons. We demonstrate that Rbpj directly activates both Notch effector genes and genes specific to mature Müller glia while indirectly repressing expression of neurogenic basic helix-loop-helix (bHLH) factors. Combined loss of function of Rbpj and Nfia/b/x resulted in conversion of nearly all Müller glia to neurons. Last, inducing Müller glial proliferation by overexpression of dominant-active Yap promotes neurogenesis in both Rbpj- and Nfia/b/x/Rbpj-deficient Müller glia. These findings demonstrate that Notch signaling and NFI factors act in parallel to inhibit neurogenic competence in mammalian Müller glia and help clarify potential strategies for regenerative therapies aimed at treating retinal dystrophies.
Figures




Update of
-
Robust reprogramming of glia into neurons by inhibition of Notch signaling and NFI factors in adult mammalian retina.bioRxiv [Preprint]. 2023 Nov 5:2023.10.29.560483. doi: 10.1101/2023.10.29.560483. bioRxiv. 2023. Update in: Sci Adv. 2024 Jul 12;10(28):eadn2091. doi: 10.1126/sciadv.adn2091. PMID: 37961663 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

