PHOSPHATASE 2A dephosphorylates PHYTOCHROME-INTERACTING FACTOR3 to modulate photomorphogenesis in Arabidopsis
- PMID: 38996075
- PMCID: PMC11449053
- DOI: 10.1093/plcell/koae200
PHOSPHATASE 2A dephosphorylates PHYTOCHROME-INTERACTING FACTOR3 to modulate photomorphogenesis in Arabidopsis
Abstract
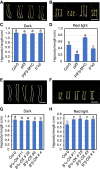
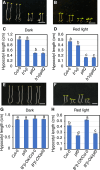
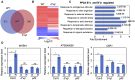
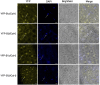
The phytochrome (phy) family of sensory photoreceptors modulates developmental programs in response to ambient light. Phys also control gene expression in part by directly interacting with the bHLH class of transcription factors, PHYTOCHROME-INTERACTING FACTORS (PIFs), and inducing their rapid phosphorylation and degradation. Several kinases have been shown to phosphorylate PIFs and promote their degradation. However, the phosphatases that dephosphorylate PIFs are less understood. In this study, we describe 4 regulatory subunits of the Arabidopsis (Arabidopsis thaliana) protein PHOSPHATASE 2A (PP2A) family (B'α, B'β, B″α, and B″β) that interact with PIF3 in yeast 2-hybrid, in vitro and in vivo assays. The pp2ab″αβ and b″αβ/b'αβ mutants display short hypocotyls, while the overexpression of the B subunits induces longer hypocotyls compared with the wild type (WT) under red light. The light-induced degradation of PIF3 is faster in the b″αβ/b'αβ quadruple mutant compared with that in the WT. Consistently, immunoprecipitated PP2A A and B subunits directly dephosphorylate PIF3-MYC in vitro. An RNA-sequencing analysis shows that B″α and B″β alter global gene expression in response to red light. PIFs (PIF1, PIF3, PIF4, and PIF5) are epistatic to these B subunits in regulating hypocotyl elongation under red light. Collectively, these data show an essential function of PP2A in dephosphorylating PIF3 to modulate photomorphogenesis in Arabidopsis.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists. All rights reserved. For commercial re-use, please contact reprints@oup.com for reprints and translation rights for reprints. All other permissions can be obtained through our RightsLink service via the Permissions link on the article page on our site—for further information please contact journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

