Risk prediction for clonal cytopenia: multicenter real-world evidence
- PMID: 38996210
- PMCID: PMC11561536
- DOI: 10.1182/blood.2024024756
Risk prediction for clonal cytopenia: multicenter real-world evidence
Abstract
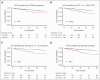
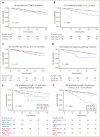
Clonal cytopenia of undetermined significance (CCUS) represents a distinct disease entity characterized by myeloid-related somatic mutations with a variant allele fraction of ≥2% in individuals with unexplained cytopenia(s) but without a myeloid neoplasm (MN). Notably, CCUS carries a risk of progressing to MN, particularly in cases featuring high-risk mutations. Understanding CCUS requires dedicated studies to elucidate its risk factors and natural history. Our analysis of 357 patients with CCUS investigated the interplay between clonality, cytopenia, and prognosis. Multivariate analysis identified 3 key adverse prognostic factors: the presence of splicing mutation(s) (score = 2 points), platelet count of <100 × 109/L (score = 2.5), and ≥2 mutations (score = 3). Variable scores were based on the coefficients from the Cox proportional hazards model. This led to the development of the clonal cytopenia risk score (CCRS), which stratified patients into low- (score of <2.5 points), intermediate- (score of 2.5 to <5), and high-risk (score of ≥5) groups. The CCRS effectively predicted 2-year cumulative incidence of MN for low- (6.4%), intermediate- (14.1%), and high-risk (37.2%) groups, respectively, by the Gray test (P < .0001). We further validated the CCRS by applying it to an independent CCUS cohort of 104 patients, demonstrating a c-index of 0.64 (P = .005) in stratifying the cumulative incidence of MN. Our study underscores the importance of integrating clinical and molecular data to assess the risk of CCUS progression, making the CCRS a valuable tool that is practical and easily calculable. These findings are clinically relevant, shaping the management strategies for CCUS and informing future clinical trial designs.
© 2024 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: R.K. reports receiving grant support from Bristol Myers Squibb (BMS); speaker bureau fees from AbbVie, Cell Therapeutics, Inc (CTI) BioPharma, Jazz Pharmaceuticals, Pharma Essentia, and Servio; and advisory board fees from AbbVie, BMS, CTI BioPharma, Geron, Jazz Pharmaceuticals, Novartis, Taiho, and Rigel Pharmaceuticals. A.P. received research funding from Pfizer and Kronos Bio; and received honoraria from BMS and AbbVie. E.A.G. has received honoraria for advisory board membership from AbbVie, Alexion Pharmaceuticals, Apellis, Celgene/BMS, CTI, BioPharma, Genentech, Novartis, PicnicHealth, Takeda Oncology, and Taiho Oncology; has received research funding from Astex Pharmaceuticals, AstraZeneca Rare Disease, Alexion Pharmaceuticals, Apellis Pharmaceuticals, Blueprint Medicines, and Genentech Inc; and received honoraria for Continuing Medical Education activities from Physicians’ Educational Resource, MediCom Worldwide, American Society of Hematology, and Aplastic Anemia and Myelodysplasia Syndrome (AAMDS) International Foundation. H.E.C. has received honoraria for advisory board memberships from AbbVie, Celgene/BMS, Genentech, Jazz Pharmaceuticals, Novartis, and Daiichi Sankyo; has received research funding from Celgene; has served on speakers bureau for BMS, Jazz Pharmaceuticals, Novartis, and Stemline Therapeutics; and has served on data safety monitoring boards for Astex, AbbVie, Takeda, and Syndax. A.M.B. received consulting or advisory board honoraria from Novartis, Acceleron, Agios, AbbVie, Takeda, Celgene/BMS, Keros Therapeutics, Taiho, and Gilead; and has research support from the National Institutes of Health Specialized Programs of Research Excellence (SPORE) in Myeloid Malignancies and the Edward P. Evans Foundation. A.M.Z. received research funding (institutional) from Celgene/BMS, AbbVie, Astex, Pfizer, MedImmune/AstraZeneca, Boehringer Ingelheim, Cardiff Oncology, Incyte, Takeda, Novartis, Aprea, and Antibody Drug Conjugates (ADC) Therapeutics; participated in advisory boards, and/or had a consultancy with, and received honoraria, from AbbVie, Otsuka, Pfizer, Celgene/BMS, Jazz Pharmaceuticals, Incyte, Agios, Boehringer Ingelheim, Novartis, Acceleron, Astellas, Daiichi Sankyo, Cardinal Health, Taiho, Seattle Genetics, BeyondSpring, Cardiff Oncology, Takeda, Ionis, Amgen, Janssen, Epizyme, Syndax, Gilead, Kura, Chiesi, ALX Oncology, BioCryst, Notable, Orum, and Tyme; and served on clinical trial committees for Novartis, AbbVie, Gilead, BioCryst, ALX Oncology, Geron, and Celgene/BMS. E.P. received honoraria from Stemline Therapeutics, Taiho, and Blueprint; and research funding from BMS, Incyte, Kura, and Syntrix Pharmaceuticals. Y.F.M. received honoraria/consulting fees from Blueprint Medicines, Geron, OncLive, and MD Education; participated in advisory boards and received honoraria from Sierra Oncology, Stemline Therapeutics, Blueprint Medicines, MorphoSys, Taiho Oncology, Rigel Pharmaceuticals, and Novartis; and received travel reimbursement from Blueprint Medicines, MD Education, and MorphoSys. J.F.Z. received honoraria from advisory boards from AbbVie, BMS, Daiichi Sankyo, Genentech, Gilead, Immunogen, Servier, and Shattuck Labs; reports consultancy for AbbVie, Foghorn, Gilead, Sellas, and Servier; and received research funding from AbbVie, Arog, Astex, Gilead, Jazz, Loxo, Merck, Newave, Shattuck Labs, Stemline Therapeutics, Sumitomo Dainippon Pharma, and Takeda. A.S. received research funding from, and serves on the advisory board for Rigel Pharmaceuticals. C.C.C. received consulting or advisory board honoraria from AbbVie, AstraZeneca, BeiGene, Genentech, MEI Pharma, TG Therapeutics, Janssen, Novartis, MingSight, Octapharma, and Lilly/Loxo; serves on an independent review committee for Octapharma; serves on steering committees for AbbVie and Lilly/Loxo; has equity in CTI Biopharma and bluebird bio; serves on speakers bureaus for AbbVie, Genentech, BeiGene, and AstraZeneca; and has received research support (to institution) from AbbVie and Lilly/Loxo. The remaining authors declare no competing financial interests.
Figures