Comparative genomics illuminates karyotype and sex chromosome evolution of sharks
- PMID: 38996479
- PMCID: PMC11406177
- DOI: 10.1016/j.xgen.2024.100607
Comparative genomics illuminates karyotype and sex chromosome evolution of sharks
Abstract
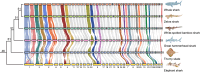
Chondrichthyes is an important lineage to reconstruct the evolutionary history of vertebrates. Here, we analyzed genome synteny for six chondrichthyan chromosome-level genomes. Our comparative analysis reveals a slow evolutionary rate of chromosomal changes, with infrequent but independent fusions observed in sharks, skates, and chimaeras. The chondrichthyan common ancestor had a proto-vertebrate-like karyotype, including the presence of 18 microchromosome pairs. The X chromosome is a conversed microchromosome shared by all sharks, suggesting a likely common origin of the sex chromosome at least 181 million years ago. We characterized the Y chromosomes of two sharks that are highly differentiated from the X except for a small young evolutionary stratum and a small pseudoautosomal region. We found that shark sex chromosomes lack global dosage compensation but that dosage-sensitive genes are locally compensated. Our study on shark chromosome evolution enhances our understanding of shark sex chromosomes and vertebrate chromosome evolution.
Keywords: cartilaginous fish; dosage compensation; microchromosome; sex chromosome; shark; vertebrate karyotype.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Yu W.-P., Rajasegaran V., Yew K., Loh W.-L., Tay B.-H., Amemiya C.T., Brenner S., Venkatesh B. Elephant shark sequence reveals unique insights into the evolutionary history of vertebrate genes: A comparative analysis of the protocadherin cluster. Proc. Natl. Acad. Sci. USA. 2008;105:3819–3824. - PMC - PubMed
-
- Kuraku S. Shark and ray genomics for disentangling their morphological diversity and vertebrate evolution. Dev. Biol. 2021;477:262–272. - PubMed
-
- Stanhope M.J., Ceres K.M., Sun Q., Wang M., Zehr J.D., Marra N.J., Wilder A.P., Zou C., Bernard A.M., Pavinski-Bitar P., et al. Genomes of endangered great hammerhead and shortfin mako sharks reveal historic population declines and high levels of inbreeding in great hammerhead. iScience. 2023;26 - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

