Transcription Attenuation in Synthetic Promoters in Nonoverlapping Tandem Formation
- PMID: 38997112
- PMCID: PMC11339919
- DOI: 10.1021/acs.biochem.4c00012
Transcription Attenuation in Synthetic Promoters in Nonoverlapping Tandem Formation
Abstract
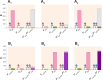
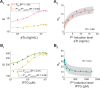
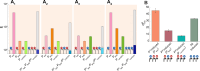
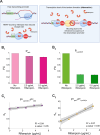
Closely spaced promoters are ubiquitous in prokaryotic and eukaryotic genomes. How their structure and dynamics relate remains unclear, particularly for tandem formations. To study their transcriptional interference, we engineered two pairs and one trio of synthetic promoters in nonoverlapping, tandem formation, in single-copy plasmids transformed into Escherichia coli cells. From in vivo measurements, we found that these promoters in tandem formation can have attenuated transcription rates. The attenuation strength can be widely fine-tuned by the promoters' positioning, natural regulatory mechanisms, and other factors, including the antibiotic rifampicin, which is known to hamper RNAP promoter escape. From this, and supported by in silico models, we concluded that the attenuation in these constructs emerges from premature terminations generated by collisions between RNAPs elongating from upstream promoters and RNAPs occupying downstream promoters. Moreover, we found that these collisions can cause one or both RNAPs to falloff. Finally, the broad spectrum of possible, externally regulated, attenuation strengths observed in our synthetic tandem promoters suggests that they could become useful as externally controllable regulators of future synthetic circuits.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

