Alcohol consumption during pregnancy differentially affects the fecal microbiota of dams and offspring
- PMID: 38997303
- PMCID: PMC11245617
- DOI: 10.1038/s41598-024-64313-z
Alcohol consumption during pregnancy differentially affects the fecal microbiota of dams and offspring
Abstract
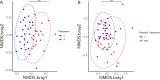
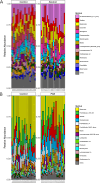
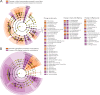
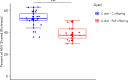
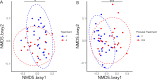
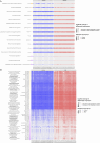
Microbiota imbalances are linked to inflammation and disease, as well as neurodevelopmental conditions where they may contribute to behavioral, physiological, and central nervous system dysfunction. By contrast, the role of the microbiota in Fetal Alcohol Spectrum Disorder (FASD), the group of neurodevelopmental conditions that can occur following prenatal alcohol exposure (PAE), has not received similar attention. Here we utilized a rodent model of alcohol consumption during pregnancy to characterize the impact of alcohol on the microbiota of dam-offspring dyads. Overall, bacterial diversity decreased in alcohol-consuming dams and community composition differed from that of controls in alcohol-consuming dams and their offspring. Bacterial taxa and predicted biochemical pathway composition were also altered with alcohol consumption/exposure; however, there was minimal overlap between the changes in dams and offspring. These findings illuminate the potential importance of the microbiota in the pathophysiology of FASD and support investigation into novel microbiota-based interventions.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

