The nature of the last universal common ancestor and its impact on the early Earth system
- PMID: 38997462
- PMCID: PMC11383801
- DOI: 10.1038/s41559-024-02461-1
The nature of the last universal common ancestor and its impact on the early Earth system
Abstract
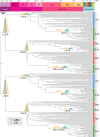
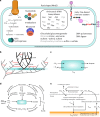
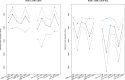
The nature of the last universal common ancestor (LUCA), its age and its impact on the Earth system have been the subject of vigorous debate across diverse disciplines, often based on disparate data and methods. Age estimates for LUCA are usually based on the fossil record, varying with every reinterpretation. The nature of LUCA's metabolism has proven equally contentious, with some attributing all core metabolisms to LUCA, whereas others reconstruct a simpler life form dependent on geochemistry. Here we infer that LUCA lived ~4.2 Ga (4.09-4.33 Ga) through divergence time analysis of pre-LUCA gene duplicates, calibrated using microbial fossils and isotope records under a new cross-bracing implementation. Phylogenetic reconciliation suggests that LUCA had a genome of at least 2.5 Mb (2.49-2.99 Mb), encoding around 2,600 proteins, comparable to modern prokaryotes. Our results suggest LUCA was a prokaryote-grade anaerobic acetogen that possessed an early immune system. Although LUCA is sometimes perceived as living in isolation, we infer LUCA to have been part of an established ecological system. The metabolism of LUCA would have provided a niche for other microbial community members and hydrogen recycling by atmospheric photochemistry could have supported a modestly productive early ecosystem.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
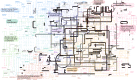
References
-
- Theobald, D. L. A formal test of the theory of universal common ancestry. Nature465, 219–222 (2010). - PubMed
-
- Woese, C. R. & Fox, G. E. The concept of cellular evolution. J. Mol. Evol.10, 1–6 (1977). - PubMed
-
- Ouzounis, C. A., Kunin, V., Darzentas, N. & Goldovsky, L. A minimal estimate for the gene content of the last universal common ancestor—exobiology from a terrestrial perspective. Res. Microbiol.157, 57–68 (2006). - PubMed
-
- Gogarten, J. P. & Deamer, D. Is LUCA a thermophilic progenote? Nat. Microbiol1, 16229 (2016). - PubMed
MeSH terms
Grants and funding
- 62220/John Templeton Foundation (JTF)
- BB/T012773/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- BB/T012951/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- GBMF9741/Gordon and Betty Moore Foundation (Gordon E. and Betty I. Moore Foundation)
- GBMF9346/Gordon and Betty Moore Foundation (Gordon E. and Betty I. Moore Foundation)
LinkOut - more resources
Full Text Sources
Miscellaneous

