A safe, effective and adaptable live-attenuated SARS-CoV-2 vaccine to reduce disease and transmission using one-to-stop genome modifications
- PMID: 38997518
- PMCID: PMC11306094
- DOI: 10.1038/s41564-024-01755-1
A safe, effective and adaptable live-attenuated SARS-CoV-2 vaccine to reduce disease and transmission using one-to-stop genome modifications
Abstract
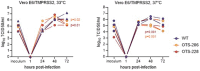
Approved vaccines are effective against severe COVID-19, but broader immunity is needed against new variants and transmission. Therefore, we developed genome-modified live-attenuated vaccines (LAV) by recoding the SARS-CoV-2 genome, including 'one-to-stop' (OTS) codons, disabling Nsp1 translational repression and removing ORF6, 7ab and 8 to boost host immune responses, as well as the spike polybasic cleavage site to optimize the safety profile. The resulting OTS-modified SARS-CoV-2 LAVs, designated as OTS-206 and OTS-228, are genetically stable and can be intranasally administered, while being adjustable and sustainable regarding the level of attenuation. OTS-228 exhibits an optimal safety profile in preclinical animal models, with no side effects or detectable transmission. A single-dose vaccination induces a sterilizing immunity in vivo against homologous WT SARS-CoV-2 challenge infection and a broad protection against Omicron BA.2, BA.5 and XBB.1.5, with reduced transmission. Finally, this promising LAV approach could be applicable to other emerging viruses.
© 2024. The Author(s).
Conflict of interest statement
Related to this work, the University of Bern has filed a patent application for the use of OTS-206 and OTS-228 as vaccine. In this application, J.S., G.T.B., B.S.T., N.J.H., A.K., L.U., F.L., J.J., N.E., D.H., M.B. and V.T. are named as inventors. The University of Bern and the Friedrich-Loeffler Institute are collaborating with RocketVax AG for the development of OTS vaccines and receive funding for research. V.T. is consulting for RocketVax AG. P.V. and V.C. are employees of RocketVax AG. The remaining authors declare no competing interests.
Figures











References
MeSH terms
Substances
Supplementary concepts
Grants and funding
- 75N92020D00005/HL/NHLBI NIH HHS/United States
- 4078P0_198473/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- 75N93022D00005/AI/NIAID NIH HHS/United States
- 75N99020D00005/OF/ORFDO NIH HHS/United States
- 22B099/Novartis Stiftung für Medizinisch-Biologische Forschung (Novartis Foundation for Medical-Biological Research)
- 31CA30_196062/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- 75N95020D00005/DA/NIDA NIH HHS/United States
- 310030B_201278/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- 75N93023D00005/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

