Identification of volatile metabolites produced from levodopa metabolism by different bacteria strains of the gut microbiome
- PMID: 38997651
- PMCID: PMC11245815
- DOI: 10.1186/s12866-024-03373-7
Identification of volatile metabolites produced from levodopa metabolism by different bacteria strains of the gut microbiome
Abstract
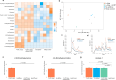
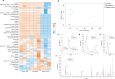
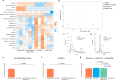
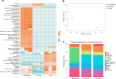
Interspecies pathways in the gut microbiome have been shown to metabolize levodopa, the primary treatment for Parkinson's disease, and reduce its bioavailability. While the enzymatic reactions have been identified, the ability to establish the resulting macromolecules as biomarkers of microbial metabolism remains technically challenging. In this study, we leveraged an untargeted mass spectrometry-based approach to investigate volatile organic compounds (VOCs) produced during levodopa metabolism by Enterococcus faecalis, Clostridium sporogenes, and Eggerthella lenta. We cultured these organisms with and without their respective bioactive metabolites and detected levodopa-induced shifts in VOC profiles. We then utilized bioinformatics to identify significant differences in 2,6-dimethylpyrazine, 4,6-dimethylpyrimidine, and 4,5-dimethylpyrimidine associated with its biotransformation. Supplementing cultures with inhibitors of levodopa-metabolizing enzymes revealed specific modulation of levodopa-associated diazines, verifying their relationship to its metabolism. Furthermore, functional group analysis depicts strain-specific VOC profiles that reflect interspecies differences in metabolic activity that can be leveraged to assess microbiome functionality in individual patients. Collectively, this work identifies previously uncharacterized metabolites of microbe-mediated levodopa metabolism to determine potential indicators of this activity and further elucidate the metabolic capabilities of different gut bacteria.
Keywords: Gut microbiota; Levodopa; Metabolism; Volatile Organoid Compound.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Relationship Between Gut Bacteria and Levodopa Metabolism.Curr Neuropharmacol. 2023;21(7):1536-1547. doi: 10.2174/1570159X21666221019115716. Curr Neuropharmacol. 2023. PMID: 36278467 Free PMC article. Review.
-
Gut bacterial deamination of residual levodopa medication for Parkinson's disease.BMC Biol. 2020 Oct 20;18(1):137. doi: 10.1186/s12915-020-00876-3. BMC Biol. 2020. PMID: 33076930 Free PMC article.
-
Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism.Science. 2019 Jun 14;364(6445):eaau6323. doi: 10.1126/science.aau6323. Science. 2019. PMID: 31196984 Free PMC article.
-
Targeting bacterial and human levodopa decarboxylases for improved drug treatment of Parkinson's disease: Discovery and characterization of new inhibitors.Eur J Pharm Sci. 2025 Aug 1;211:107133. doi: 10.1016/j.ejps.2025.107133. Epub 2025 May 20. Eur J Pharm Sci. 2025. PMID: 40403986
-
Gut Microbiome and Response to Cardiovascular Drugs.Circ Genom Precis Med. 2019 Sep;12(9):421-429. doi: 10.1161/CIRCGEN.119.002314. Epub 2019 Aug 28. Circ Genom Precis Med. 2019. PMID: 31462078 Free PMC article. Review.
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

