Effects of Low-Salinity Stress on Histology and Metabolomics in the Intestine of Fenneropenaeus chinensis
- PMID: 38997992
- PMCID: PMC11240639
- DOI: 10.3390/ani14131880
Effects of Low-Salinity Stress on Histology and Metabolomics in the Intestine of Fenneropenaeus chinensis
Abstract
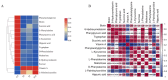
Metabolomics has been used extensively to identify crucial molecules and biochemical effects induced by environmental factors. To understand the effects of acute low-salinity stress on Fenneropenaeus chinensis, intestinal histological examination and untargeted metabonomic analysis of F. chinensis were performed after exposure to a salinity of 15 ppt for 3, 7, and 14 d. The histological examination revealed that acute stress resulted in most epithelial cells rupturing, leading to the dispersion of nuclei in the intestinal lumen after 14 days. Metabolomics analysis identified numerous differentially expressed metabolites (DEMs) at different time points after exposure to low-salinity stress, in which some DEMs were steadily downregulated at the early stage of stress and then gradually upregulated. We further screened 14 overlapping DEMs, in which other DEMs decreased significantly during low-salinity stress, apart from L-palmitoylcarnitine and vitamin A, with enrichments in phenylalanine, tyrosine and tryptophan biosynthesis, fatty acid and retinol metabolism, and ABC transporters. ABC transporters exhibit significant abnormalities and play a vital role in low-salinity stress. This study provides valuable insights into the molecular mechanisms underlying the responses of F. chinensis to acute salinity stress.
Keywords: ABC transporters; metabolites; morphology; salinity; shrimp intestine.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Li Z., Li J., Wang Q., He Y., Liu P. Aflp-Based Genetic Linkage Map of Marine Shrimp Penaeus (Fenneropenaeus chinensis) Aquaculture. 2006;261:463–472. doi: 10.1016/j.aquaculture.2006.07.002. - DOI
-
- Liu J., Zhang D., Zhang L., Wang Z., Shen J. New Insight on Vitality Differences for the Penaeid Shrimp, Fenneropenaeus chinensis, in Low Salinity Environment Through Transcriptomics. Front. Ecol. Evol. 2022;10:716018. doi: 10.3389/fevo.2022.716018. - DOI
-
- Gao W., Tian L., Huang T., Yao M., Hu W., Xu Q. Effect of Salinity on the Growth Performance, Osmolarity and Metabolism-Related Gene Expression in White Shrimp Litopenaeus vannamei. Aquac. Rep. 2016;4:125–129. doi: 10.1016/j.aqrep.2016.09.001. - DOI
-
- Pan L.Q., Luan Z.H., Jin C.X. Effects of Na+/K+ and Mg2+/Ca2+ Ratios in Saline Groundwaters On Na+–K+-Atpase Activity, Survival and Growth of Marsupenaeus Japonicus Postlarvae. Aquaculture. 2006;261:1396–1402. doi: 10.1016/j.aquaculture.2006.09.031. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

