Genome-Wide Identification and Characterization of RdHSP Genes Related to High Temperature in Rhododendron delavayi
- PMID: 38999718
- PMCID: PMC11244423
- DOI: 10.3390/plants13131878
Genome-Wide Identification and Characterization of RdHSP Genes Related to High Temperature in Rhododendron delavayi
Abstract
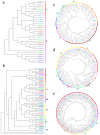
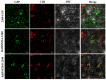
Heat shock proteins (HSPs) are molecular chaperones that play essential roles in plant development and in response to various environmental stresses. Understanding R. delavayi HSP genes is of great importance since R. delavayi is severely affected by heat stress. In the present study, a total of 76 RdHSP genes were identified in the R. delavayi genome, which were divided into five subfamilies based on molecular weight and domain composition. Analyses of the chromosome distribution, gene structure, and conserved motif of the RdHSP family genes were conducted using bioinformatics analysis methods. Gene duplication analysis showed that 15 and 8 RdHSP genes were obtained and retained from the WGD/segmental duplication and tandem duplication, respectively. Cis-element analysis revealed the importance of RdHSP genes in plant adaptations to the environment. Moreover, the expression patterns of RdHSP family genes were investigated in R. delavayi treated with high temperature based on our RNA-seq data, which were further verified by qRT-PCR. Further analysis revealed that nine candidate genes, including six RdHSP20 subfamily genes (RdHSP20.4, RdHSP20.8, RdHSP20.6, RdHSP20.3, RdHSP20.10, and RdHSP20.15) and three RdHSP70 subfamily genes (RdHSP70.15, RdHSP70.21, and RdHSP70.16), might be involved in enhancing the heat stress tolerance. The subcellular localization of two candidate RdHSP genes (RdHSP20.8 and RdHSP20.6) showed that two candidate RdHSPs were expressed and function in the chloroplast and nucleus, respectively. These results provide a basis for the functional characterization of HSP genes and investigations on the molecular mechanisms of heat stress response in R. delavayi.
Keywords: RNA-seq data; RdHSP gene family; expression pattern; high-temperature stress; subcellular localization.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures









References
-
- Francini A., Sebastiani L. Abiotic Stress Effects on Performance of Horticultural Crops. Horticulturae. 2019;5:67. doi: 10.3390/horticulturae5040067. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

