Insights into the Clinical, Biological and Therapeutic Impact of Copy Number Alteration in Cancer
- PMID: 38999925
- PMCID: PMC11241182
- DOI: 10.3390/ijms25136815
Insights into the Clinical, Biological and Therapeutic Impact of Copy Number Alteration in Cancer
Abstract
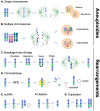
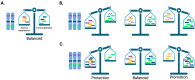
Copy number alterations (CNAs), resulting from the gain or loss of genetic material from as little as 50 base pairs or as big as entire chromosome(s), have been associated with many congenital diseases, de novo syndromes and cancer. It is established that CNAs disturb the dosage of genomic regions including enhancers/promoters, long non-coding RNA and gene(s) among others, ultimately leading to an altered balance of key cellular functions. In cancer, CNAs have been associated with almost all steps of the disease: predisposition, initiation, development, maintenance, response to treatment, resistance, and relapse. Therefore, understanding how specific CNAs contribute to tumourigenesis may provide prognostic insight and ultimately lead to the development of new therapeutic approaches to improve patient outcomes. In this review, we provide a snapshot of what is currently known about CNAs and cancer, incorporating topics regarding their detection, clinical impact, origin, and nature, and discuss the integration of innovative genetic engineering strategies, to highlight the potential for targeting CNAs using novel, dosage-sensitive and less toxic therapies for CNA-driven cancer.
Keywords: cancer; genetic; prognostic; targeted therapy.
Conflict of interest statement
R.S.K. discloses advisory board participation from Jazz Pharmaceuticals, Amgen and Link Healthcare. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Hierarchical discovery of large-scale and focal copy number alterations in low-coverage cancer genomes.BMC Bioinformatics. 2020 Apr 16;21(1):147. doi: 10.1186/s12859-020-3480-3. BMC Bioinformatics. 2020. PMID: 32299346 Free PMC article.
-
Somatic Copy Number Alterations and Associated Genes in Clear-Cell Renal-Cell Carcinoma in Brazilian Patients.Int J Mol Sci. 2021 Feb 25;22(5):2265. doi: 10.3390/ijms22052265. Int J Mol Sci. 2021. PMID: 33668731 Free PMC article.
-
Genomic copy number alterations as biomarkers for triple negative pregnancy-associated breast cancer.Cell Oncol (Dordr). 2022 Aug;45(4):591-600. doi: 10.1007/s13402-022-00685-6. Epub 2022 Jul 6. Cell Oncol (Dordr). 2022. PMID: 35792986 Free PMC article.
-
Methods for copy number aberration detection from single-cell DNA-sequencing data.Genome Biol. 2020 Aug 17;21(1):208. doi: 10.1186/s13059-020-02119-8. Genome Biol. 2020. PMID: 32807205 Free PMC article. Review.
-
Copy number alteration signatures as biomarkers in cancer: a review.Biomark Med. 2022 Apr;16(5):371-386. doi: 10.2217/bmm-2021-0476. Epub 2022 Feb 23. Biomark Med. 2022. PMID: 35195030 Review.
Cited by
-
The Histomorphology to Molecular Transition: Exploring the Genomic Landscape of Poorly Differentiated Epithelial Endometrial Cancers.Cells. 2025 Mar 5;14(5):382. doi: 10.3390/cells14050382. Cells. 2025. PMID: 40072110 Free PMC article. Review.
-
Replication stress increases de novo CNVs across the malaria parasite genome.bioRxiv [Preprint]. 2024 Dec 31:2024.12.19.629492. doi: 10.1101/2024.12.19.629492. bioRxiv. 2024. PMID: 39803504 Free PMC article. Preprint.
-
Genetic background and oncogenic driver determines the genomic evolution and transcriptomics of mammary tumor metastasis.Commun Biol. 2025 Aug 14;8(1):1224. doi: 10.1038/s42003-025-08624-5. Commun Biol. 2025. PMID: 40813609 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

