Identification of New Chemoresistance-Associated Genes in Triple-Negative Breast Cancer by Single-Cell Transcriptomic Analysis
- PMID: 38999963
- PMCID: PMC11241600
- DOI: 10.3390/ijms25136853
Identification of New Chemoresistance-Associated Genes in Triple-Negative Breast Cancer by Single-Cell Transcriptomic Analysis
Abstract
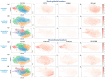
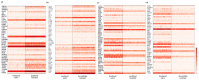
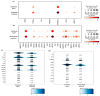
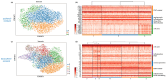
Triple-negative breast cancer (TNBC) is a particularly aggressive mammary neoplasia with a high fatality rate, mainly because of the development of resistance to administered chemotherapy, the standard treatment for this disease. In this study, we employ both bulk RNA-sequencing and single-cell RNA-sequencing (scRNA-seq) to investigate the transcriptional landscape of TNBC cells cultured in two-dimensional monolayers or three-dimensional spheroids, before and after developing resistance to the chemotherapeutic agents paclitaxel and doxorubicin. Our findings reveal significant transcriptional heterogeneity within the TNBC cell populations, with the scRNA-seq identifying rare subsets of cells that express resistance-associated genes not detected by the bulk RNA-seq. Furthermore, we observe a partial shift towards a highly mesenchymal phenotype in chemoresistant cells, suggesting the epithelial-to-mesenchymal transition (EMT) as a prevalent mechanism of resistance in subgroups of these cells. These insights highlight potential therapeutic targets, such as the PDGF signaling pathway mediating EMT, which could be exploited in this setting. Our study underscores the importance of single-cell approaches in understanding tumor heterogeneity and developing more effective, personalized treatment strategies to overcome chemoresistance in TNBC.
Keywords: 3D spheroids; bulk RNA-sequencing; chemoresistance; single-cell RNA-sequencing; transcriptomics; triple-negative breast cancer.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

